温馨提示:请使用火狐或者Chrome的网页浏览器来查看报告
微科盟动植物基因组重测序分析结题报告
一 概述
重测序(Resequencing)是指在已有参考基因组的情况下,对目标个体或者群体再度进行测序,以获得其在靶向基因区域或者全基因组上的变异情况。这些变异信息可进一步用于研究物种遗传多样性、进化关系与基因功能变化等。重测序技术已广泛应用于基础研究、临床诊断和分子育种等领域。
本次测序采用Illumina测序平台。原始数据在测序后进行质控,以去除低质量数据。高质量的测序数据将与参考基因组进行比对,并进一步用于单核苷酸多态性位点(Single Nucleotide Polymorphism, SNP)、小片段插入/缺失(Insertion/Deletion, InDel)、结构性变异(Structural Variation, SV)、拷贝数变异(Copy Number Variation, CNV)上的变异分析。
二 项目流程
2.1 测序实验流程
从DNA样品提取到最终数据获得,样品检测、建库、测序等每一环节都会直接影响数据的数量和质量,从而影响后续信息分析的结果。为从源头保证测序数据准确可靠,我们承诺在数据的所有生产环节都严格把关,从根源上确保高质量数据的产出。建库测序的流程图如下(图2.1):
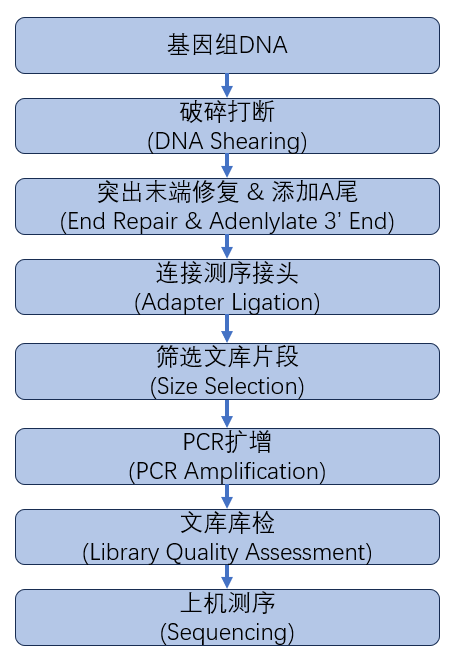
图2.1 DNA建库测序流程
2.1.2 文库构建与质检
文库构建
文库构建原理如图2.2所示。
- 破碎、补齐末端:在构建文库时,首先将基因组DNA经Covaris破碎仪随机打断成长度为350bp左右的片段。因片段化后的DNA存在5’或3’端突出,不方便引物接入,所以需要补齐纯化后的DNA片段的末端。本次建库使用T4 DNA 聚合酶(T4 DNA Polymerase)的外切酶(Exonuclease)活性消化3’端的单链突出,并使用其聚合酶(Polymerase)活性补齐5’端的突出。在修补末端后,使用磷酸激酶(PNK)在5’末端加上后续连接反应必需的磷酸基团,再经过Agencourt AMpure XP 磁珠纯化,最终得到5’端含有磷酸基团的平末端DNA短片段文库。
- 连接测序接头:在获得平末端DNA短片段文库后,向末端修饰完成的双链DNA 3’末端加上单个腺苷酸“A”,以防止DNA片段之间的平末端自我连接。然后加入连接缓冲液和双链测序接头,利用T4 DNA 连接酶将Illumina测序接头连接至文库DNA 两端。
- 文库片段筛选:文库加入接头后,使用Agencourt SPRIselect 核酸片段筛选试剂盒纯化文库,并使用两步法筛选(Double Size selection),先用SPRI磁珠去掉目标域左侧小片段(Left-side Size selection),再去掉位于目标片段区域右侧的大片段(Right-side Size selection)。最终筛选出片段长度适中的原始文库,用于下一步的PCR扩增。
- PCR扩增:扩增中使用高保真的聚合酶扩增原始文库,以保证文库总量足够用于上机。由于只有两端都连有接头的DNA片段才能被扩增,因此该步骤还能够有效富集目标DNA,减少因扩增循环数过大而引入的bias。
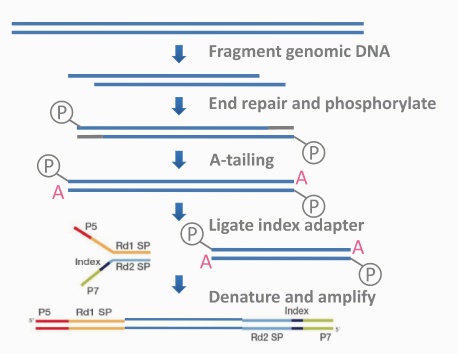
图2.2 文库构建原理示意图
测序接头:包括P5/P7,index和Rd1/Rd2 SP三个部分(如上图所示)。其中P5/P7是PCR扩增引物及flow cell上引物结合的部分,index提供区分不同文库的信息,Rd1/Rd2 SP即read1/read2 sequence primer,是测序引物结合区域,测序过程理论上由Rd1/Rd2 SP向后开始进行。
文库质检
扩增结束后,使用Qubit3.0 精确测定每个文库浓度。随后使用Agilent 5400 对文库的Insert size 进行检测,Insert size 符合预期后,使用qPCR 方法对文库的有效浓度(>1.5 nM)进行准确定量,以保证文库质量。
2.1.3 上机测序
库检合格后,把不同文库按照有效浓度及目标下机数据量的需求pooling后进行Illumina测序。测序的基本原理是边合成边测序(Sequencing by Synthesis)。在测序的flow cell中加入四种荧光标记的dNTP、DNA聚合酶以及接头引物进行扩增,在每一个测序簇延伸互补链时,每加入一个被荧光标记的dNTP就能释放出相对应的荧光,测序仪通过捕获荧光信号,并通过计算机软件将光信号转化为测序峰,从而获得待测片段的序列信息。测序过程如下图所示(图2.3):

图2.3 Illumina测序原理示意图
2.2 生信分析流程
重测序的主要目的是获取样本中的变异信息。在将样本序列比对至参考基因组后,可以依据变异类型的不同,使用不同的统计模型检测出可靠的变异情况。本次检测的变异类型包括单核苷酸多态性(SNP)、小片段插入/缺失(InDel)、结构性变异(SV)与拷贝数变异(SNV)。信息分析流程如下图所示:
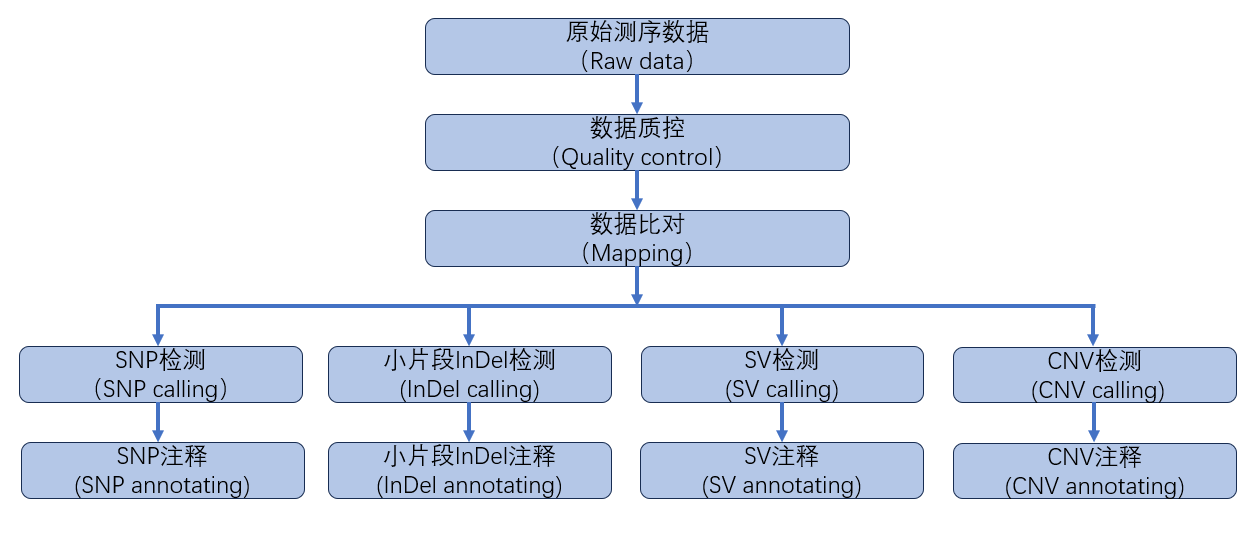
图2.4 重测序信息分析技术流程
三 结果文件解读
3.1 数据质量控制
流程结果
├── 01.QA (进入文件目录)
│ ├── Sample1
│ │ ├── 图3.1.1_过滤前碱基质量分布_Sample1.svg
│ │ ├── 图3.1.2_过滤后碱基质量分布_Sample1.svg
│ │ ├── 图3.1.3_过滤前GC含量分布_Sample1.svg
│ │ ├── 图3.1.4_过滤后GC含量分布_Sample1.svg
│ │ └── 图3.1.5_过滤reads比例_Sample1.svg
│ ├── ...
│ ├── 表3.1.1_各样本质控前质量情况汇总.csv
│ └── 表3.1.2_各样本质控后质量情况汇总.csv
└── ...
用fastp软件[1]对每一个样本的测序数据raw data做质控处理(参数: --overrepresentation_analysis --trim_front1 2 --trim_front2 2 --cut_front --cut_tail --cut_window_size 3 --cut_mean_quality 30 ),得到clean data用于下游分析,质控规则如下:
- 对整条read做保留或弃去处理
- 根据碱基质量值弃去低质量的整条read
- 当一条read中N碱基的个数达到5个时,弃去整一条read;
- 当一条read中低质量碱基(此处低质量阈值为15)达到40%时,弃去整一条read。
- 根据read长度弃去低质量的整条read
- 当一条read的碱基个数少于15时,弃去整一条read。
- 对read做局部裁剪
- 强制性删除头部2个碱基与尾部2个碱基
- 因为测序反应一般起始和结束阶段的碱基质量是最差的,所以我们统一裁剪掉read1和read2它们各自的5'端的2个碱基、3'端的2个碱基。
- 根据碱基质量值删除滑动窗口内的碱基
- 一般来说,测出来的read,开头的碱基质量差,然后随着测序反应的稳定,碱基质量会逐步上升,后面随着反应时间的延长、化学产物的积累、酶受到的环境不利影响的积累,到后面碱基质量会下降,所以需要对read的开头和结尾的低质量碱基区做裁剪。
- 我们以3个碱基的长度为一个窗口的大小,从read的头部向尾部滑动,计算窗口内碱基的平均质量值。如果平均质量值小于阈值30,则表明当前窗口的碱基质量差,随后删除这个窗口内的所有碱基,然后滑动到下一个窗口。上述操作将重复至在该方向上发现平均质量高于质量阈值的窗口为止。同理,从read的尾部向头部,也做类似的滑动窗口删除碱基的处理。
- adapter序列裁剪
- 默认情况下,对pair-end 的read1和read2做两序列比对,查看比对结果中,read1的右端是否会超出read2的右端,read2的左端是否会超出read1的左端,如果超出的话,表明是测到了adapter序列,则会把超出的部分给剪掉。
将所有样本的质控信息做提取整合,生成总体的样本测序数据质控信息统计表。表格右上角的检索框可用于筛选包含检索内容的条目,即仅显示包含输入关键词的行
| Sample ID | Total reads | Total bases | Q20 bases | Q30 bases | Q20 rate | Q30 rate | Read1 mean length | Read2 mean length | Gc content |
|---|---|---|---|---|---|---|---|---|---|
| Sample1 | 24127002 | 3619050300 | 3504891174 | 3323230925 | 0.968456 | 0.918260 | 150 | 150 | 0.378251 |
| Sample2 | 23991428 | 3598714200 | 3496304045 | 3328742699 | 0.971543 | 0.924981 | 150 | 150 | 0.379382 |
| Sample3 | 23882216 | 3582332400 | 3469634147 | 3291120449 | 0.968541 | 0.918709 | 150 | 150 | 0.384092 |
| Sample4 | 23390452 | 3508567800 | 3395002339 | 3215827031 | 0.967632 | 0.916564 | 150 | 150 | 0.381878 |
| Sample5 | 23977090 | 3596563500 | 3484329904 | 3306443786 | 0.968794 | 0.919334 | 150 | 150 | 0.382440 |
表3.1.1 各样本质控前质量情况汇总
| Sample ID | Total reads | Total bases | Q20 bases | Q30 bases | Q20 rate | Q30 rate | Read1 mean length | Read2 mean length | Gc content |
|---|---|---|---|---|---|---|---|---|---|
| Sample1 | 23967838 | 3445130871 | 3355531122 | 3187518038 | 0.973992 | 0.925224 | 143 | 143 | 0.378200 |
| Sample2 | 23860312 | 3432629154 | 3351763952 | 3196247975 | 0.976442 | 0.931137 | 143 | 143 | 0.379382 |
| Sample3 | 23724572 | 3408549762 | 3320587738 | 3155682387 | 0.974194 | 0.925814 | 143 | 143 | 0.383954 |
| Sample4 | 23237050 | 3339619115 | 3250170142 | 3084346332 | 0.973216 | 0.923562 | 143 | 143 | 0.381771 |
| Sample5 | 23819552 | 3425421261 | 3337246640 | 3172618571 | 0.974259 | 0.926198 | 144 | 143 | 0.382319 |
表3.1.2 各样本质控后质量情况汇总
- sample id: 样本ID
- total reads: 样本序列总数
- total bases: 样本碱基总数
- total giga bases: 样本碱基总数(G为单位)
- q20 bases: 质量分高于20(错误率0.01)的碱基总数
- q30 bases: 质量分高于30(错误率0.001)的碱基总数
- q20 rate: 质量分高于20(错误率0.01)的碱基占比(Q20)
- q30 rate: 质量分高于30(错误率0.001)的碱基占比(Q30)
- read1 mean length: read1平均长度
- read2 mean length: read2平均长度
- gc content: GC含量
- 图3.1.1与图3.1.2展示了过滤前后每个reads在对应碱基上的平均质量, 其中绿色区域表示质量优秀,黄色区域表示合格,红色区域表示不合格。
- 图3.1.3与图3.1.4展示了过滤前后每个reads在对应碱基上的GC含量。 不同物种的GC含量不同,但在同一物种的测序reads上,GC含量应该相对稳定。
- 图3.1.5展示了每个样本中,因各种原因被过滤掉的reads的比例。
3.2 比对参考基因组
流程结果
├── 02.mapping (进入文件目录)
│ ├── Sample1
│ │ ├── 图3.2.1_各染色体平均覆盖率_Sample1.svg
│ │ ├── 图3.2.2_样本质量分布_Sample1.svg
│ │ └── 图3.2.3_样本基因组覆盖率分布_Sample1.svg
│ ├── ...
│ ├── 表3.2.1_原始参考基因组统计数据.csv
│ ├── 表3.2.2_过滤后参考基因组统计数据.csv
│ └── 表3.2.3_去重后各样本比对结果.csv
└── ...
用bwa软件的mem算法[2]将每一个样本的测序数据raw data比对至参考基因组上(参数: -A 1 -B 4 -E 1 -O 6 -T 30 -d 100 )。得到比对文件经过sambamba markdup去重后用于下游分析。
BWA MEM算法及参数解释如下:
- BWA-MEM使用种子序列定位及延伸算法(seed and extend)进行比对。 在该算法下,程序会首先查找reads中的片段于参考基因组中的最长精确匹配(即种子), 然后尝试沿参考基因组扩展这些匹配以延伸种子序列。经过评分与动态规划后,保留最优的比对结果。 这种方法可以提高比对的速度和准确性,适用于100bp-1Mp的reads。
- 该算法的计算步骤如下
- 生成种子:寻找reads中的小片段与参考基因组的最长精确匹配(种子),这一段序列将作为比对的起点
- 链构建:将共线且彼此相邻的种子连接后形成链(chain)。
- 种子排序:基于种子所处的链长度将种子排序。已经具有最佳比对的种子将会从这一动态过程中被排除。
- 种子延伸:将序号较高的种子向左右延伸,并计算每个位点的延伸得分。延伸过程中的最高得分(即局部最高得分)将被记录。
- 输出最终结果:若当前延伸得分与延伸过程中的局部最高得分的差值高于预设值1时,终止延伸。 此时进一步将这一差值与另一预设值2比较。若差值高于预设值2,则输出局部最高得分时的延伸序列;若低于预设值2,则输出当前延伸序列。
- BWA MEM的运行参数解释如下
- -A match_score: 延伸时,reads上的下一个碱基匹配至参考基因组上后增加的分数(默认为1,当前为1)。
- -B mismatch_penalty: 延伸时,reads上的下一个碱基未能匹配至参考基因组上后减少的分数(默认为4,当前为4)。
- -E extend_penalty: 延伸时,每增加一个gap所减少的分数(默认为1,当前为1)。
-
-O gap_penalty: 延伸时,创建gap时的基底罚分(默认为6,当前为6)。
- 该参数与-E一起组成单项gap的总罚分,公式为:O + k*E, 其中k为gap的长度。
- -T mapq_threshold: 即预设值2,用于输出最终结果时的判断(默认为30,当前为30)。
- -d z_dropoff: 即预设值1,用于判断是否终止延伸(默认为100,当前为100)。
本次比对中使用的参考基因组为GCF_018340385_1_ASM1834038v1_genomic,物种为Cyprinus_carpio,统计信息如下:
| Seq numbers | Total length | Q1 | Q2 | Q3 | Mean Length | GC content(%) | N50 | L50 | N90 | L90 |
|---|---|---|---|---|---|---|---|---|---|---|
| 6701 | 1680134903 | 1786.0 | 1980.0 | 3942.0 | 250729.0 | 36.93 | 29545497 | 24 | 20763676 | 50 |
表3.2.1 原始参考基因组统计数据
在过滤长度低于2000bp的序列后,统计信息如下
| Seq numbers | Total length | Q1 | Q2 | Q3 | Mean Length | GC content(%) | N50 | L50 | N90 | L90 |
|---|---|---|---|---|---|---|---|---|---|---|
| 3247 | 1674466193 | 2304.5 | 4167.0 | 14116.0 | 515696.4 | 36.92 | 29545497 | 24 | 20928587 | 49 |
表3.2.2 过滤后参考基因组统计数据
- Seq numbers: 基因组中序列总数
- Total length: 基因组总碱基数
- Q1: 基因组长度的下四分位数
- Q2: 基因组长度的中位数
- Q3: 基因组长度的上四分位数
- Mean Length: 基因组长度的平均数
- GC content(%): 基因组中GC含量占比
- N50: 基因组长度排序后,从长到短累加总值达到基因组总长度50%时的序列长度
- L50: 基因组长度排序后,从长到短累加总值达到基因组总长度50%时的序列序号
- N90: 同N50,但累加长度占比为90%
- L90: 同L90,但累加长度占比为90%
| Sample ID | Total reads | Number of mapped reads | Number of mapped bases | Number of duplicated reads | Error rate | Mean coverage | Coverage >= 1X | Coverage >= 4X |
|---|---|---|---|---|---|---|---|---|
| Sample1 | 24666606 | 24519811 | 3340056365 | 1186468 | 0.02 | 1.99 | 67.60 | 17.69 |
| Sample2 | 24562246 | 24424606 | 3329519373 | 1206873 | 0.02 | 1.99 | 67.08 | 17.66 |
| Sample3 | 24407068 | 24263026 | 3307692281 | 1212787 | 0.02 | 1.98 | 67.13 | 17.28 |
| Sample4 | 23910279 | 23763278 | 3238383028 | 1179109 | 0.02 | 1.93 | 67.08 | 16.66 |
| Sample5 | 24510973 | 24366177 | 3322796205 | 1267291 | 0.02 | 1.98 | 67.25 | 17.50 |
表3.2.3 去重后各样本比对结果
- sample id: 样本ID
- Total reads: 样本序列总数
- Number of mapped reads: 比对至参考基因组上的序列总数
- Number of mapped bases: 比对至参考基因组上的碱基总数
- Number of duplicated reads: 被标记为重复的序列总数
- Error rate: 比对错误率
- Mean Depth: 平均覆盖率
- Coverage >= 1X: 覆盖率大于等于1的碱基占比
- Coverage >= 2X: 覆盖率大于等于2*物种单倍体数的碱基占比
- 图3.2.1展示了每个contig上的平均覆盖率,其中横坐标为contig的序号,纵坐标为平均覆盖率。 由于每个参考基因组中contig数量不同且部分参考基因组会有较多contig,此处只展示前25个contig的情况。
- 图3.2.2展示了每个样本的比对质量分布,其中横坐标为比对质量,纵坐标为比对质量的频数。
- 图3.2.3展示了每个样本的覆盖率分布,其中横坐标为覆盖率, 纵坐标为参考基因组有多少百分比的碱基具有该覆盖率。
3.3 SNP检测与注释
流程结果
├── 03.SNP (进入文件目录)
│ ├── Sample1
│ │ ├── 表3.3.5_SNP密度统计_Sample1.csv (因数据过多,未列入报告)
│ │ ├── 图3.3.1_样本SNP效应类型占比_Sample1.png
│ │ ├── 图3.3.2_样本SNP发生位点占比_Sample1.png
│ │ └── 图3.3.4_样本SNP密度热力图_Sample1.png
│ ├── ...
│ ├── 表3.3.2_SNP变异信息统计(按对基因的影响预测统计).csv
│ ├── 表3.3.3_SNP变异信息统计(按区域统计).csv
│ ├── 表3.3.4_SNP碱基变化统计.csv
│ └── 图3.3.3_样本SNP碱基变化堆叠柱形图.png
└── ...
SNP位点指的是单核苷酸多态性位点(Single Nucleotide Polymorphism),即基因组上由单个核苷酸变异形成的遗传标记,在研究群体系谱以及遗传差异时具有重要的指示作用。 本次分析用Freebayes软件[3]对所有样本的比对数据(bam)进行变异位点分析(关键参数: --theta 0.001 --min-mapping-quality 30 --min-base-quality 20 --mismatch-base-quality-threshold 10 --read-max-mismatch-fraction 1.0 --min-coverage 0 --read-dependence-factor 0.9 --posterior-integration-limits ['1', '3'] )。在过滤掉可信度低的变异后,使用SnpEff软件[4]对变异位点进行注释。
Freebayes的算法及参数介绍如下:
- Freebayes基于Bayesian模型,估算在给定的预期等位基因频率、样本多样性与测序随机误差下,提供的所有样本中存在变异的概率。 其模型框架对复杂的变异类型也具有较高的识别精度,并支持多样本同时分析,从而更精确地计算群体中的遗传组成。由于不依赖于参考数据库或已知变异的先验信息,这一算法能对更多物种/变异情况进行检验。
- 该算法的计算步骤如下
- 生成pile-up图:将比对文件中的每一条记录将按照其在参考基因组上的位置进行排列,这一过程中会有多条reads沿参考基因组发生堆叠,所以称为pile-up。
- 寻找候选变异:检测参考基因组上每个位点/区域可能出现的基因型组合(包括reads与参考基因组)。如果大多数reads在某一位点上与参考基因组拥有不同的碱基,那么这个位点将被记录为SNP候选位点。
- 变异校验:对于每个候选变异,计算给定基因型组合下出现reads中变异的似然概率,并通过似然概率,计算给定reads中出现给定基因型组合的后验概率。
- 输出最终结果:后验概率最高的位点将被作为最终结果输出,其可信度将被转化为QUAL分数记录在变异文件内。
- Freebayes的运行参数解释如下
- --theta: 即先验的等位基因变异频率(默认为0.001,当前为0.001)。此值会影响Freebayes如何权衡零假设(没有变异)和备择假设(有变异)之间的权重。
- --min-mapping-quality: 考虑变异时,位点所处reads的最低比对质量(默认为30,当前为30)
- --min-base-quality: 考虑变异时,证据碱基的最低测序质量(默认为20,当前为20)
- --mismatch-base-quality-threshold: 考虑比对位点为错配时的碱基测序质量阈值(默认为20,当前为10)
- --read-max-mismatch-fraction: Reads中合格的mismatch位点占比阈值(默认为1,当前为1.0)
- --min-coverage: 考虑候选变异位点时的最低覆盖率(默认为10,当前为0)
- --read-dependence-factor: 不同reads之间对应位点来源于同一祖先的可能性,即相关度(默认为0.9,当前为0.9)
- --posterior-integration-limits N,M: 调整后验概率的计算模式。在计算后验概率时,将对所有样本的似然概率进行排序,并取前N个样本参与计算。 对于上述N个样本中的每一个,只有前M个局部最佳的数据似然性参与计算(默认为[1,3],当前为['1', '3'])
在检测完成后,需要对变异位点进行过滤,以保证结果的可信度。本次分析中变异位点的过滤规则如下:
- 保留QUAL分数大于30的位点(推荐值为:30)
- QUAL分数表示变异检测的质量。其数值由假阳性概率以10为底的负对数表示,即 QUAL= -10 * log10(假阳性概率)。QUAL分数越高,说明变异检测越可信。
- 保留SAF > 0 且 SAR > 0的位点(推荐值为:SAF > 0 & SAR > 0)
- SAF表示正链中支持该变异的证据reads数,SAR表示反链中支持该变异的证据reads数
- 保留RPR > 1 且 RPL > 1的位点(推荐值为:RPR > 1 & RPL > 1)
- RPR表示变异位点3'端证据reads数,RPL表示变异位点5'端证据reads数
- 保留DP > 1的位点(推荐值为:DP > 1)
- DP表示变异位点的深度,即该位点上的reads数
- 保留 0.25 <= AB <= 0.75的位点(推荐值为:0.25 <= AB <= 0.75)
- AB表示杂合位点上,与参考基因组持有相同碱基的reads数占总证据reads的比例。对于真实的杂合位点,一般期望AB的值在0.5左右。过于极端的数值可能是由测序偏差等因素产生
- 保留 0.9 <= MQM / MQMR <= 1.05的位点(推荐值为:0.9 <= MQM / MQMR <= 1.05)
- MQM表示支持该变异的证据reads在该位点上的平均比对质量,MQMR表示支持参考基因组的证据reads在该位点上的平均比对质量。如果没有技术偏差,这二类reads的比对质量应相近
- 保留 F_MISSING < 0.1 的位点(推荐值为:F_MISSING < 0.1)
- F_MISSING表示该位点上缺失的样本数占总样本数的比例。
以下为结果文件示例
| #CHROM | POS | ID | REF | ALT | QUAL | FILTER | INFO | FORMAT | Sample1 |
|---|---|---|---|---|---|---|---|---|---|
| NC_056572.1 | 25436 | . | C | T | 191.584 | PASS | AB=0.5;ABP=3.0103;AC=2;AF=0.7;AN=2;AO=8;CIGAR=1X;DP=11;DPB=11;DPRA=1.125;EPP=3.0103;EPPR=3.73412;GTI=1;LEN=1;MEANALT=1;MQM=60;MQMR=60;NS=5;NUMALT=1;ODDS=2.7786;PAIRED=1;PAIREDR=1;PAO=0;PQA=0;PQR=0;PRO=0;QA=296;QR=111;RO=3;RPL=5;RPP=4.09604;RPPR=3.73412;RPR=3;RUN=1;SAF=3;SAP=4.09604;SAR=5;SRF=2;SRP=3.73412;SRR=1;TYPE=snp;technology.illumina=1;ANN=T|3_prime_UTR_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771354.1|protein_coding|13/13|c.*13G>A|||||13|,T|3_prime_UTR_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771433.1|protein_coding|13/13|c.*13G>A|||||13|,T|3_prime_UTR_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771494.1|protein_coding|13/13|c.*13G>A|||||13| | GT:DP:AD:RO:QR:AO:QA:GL | 1/1:3:0,3:0:0:3:111:-10.3539,-0.90309,0 |
| NC_056572.1 | 25512 | . | C | G | 237.982 | PASS | AB=0.5;ABP=3.0103;AC=2;AF=0.9;AN=2;AO=8;CIGAR=1X;DP=9;DPB=9;DPRA=0;EPP=4.09604;EPPR=5.18177;GTI=0;LEN=1;MEANALT=1;MQM=60;MQMR=60;NS=5;NUMALT=1;ODDS=5.00439;PAIRED=1;PAIREDR=1;PAO=0;PQA=0;PQR=0;PRO=0;QA=296;QR=37;RO=1;RPL=4;RPP=3.0103;RPPR=5.18177;RPR=4;RUN=1;SAF=5;SAP=4.09604;SAR=3;SRF=0;SRP=5.18177;SRR=1;TYPE=snp;technology.illumina=1;ANN=G|synonymous_variant|LOW|LOC109102258|LOC109102258|transcript|XM_042771354.1|protein_coding|13/13|c.1620G>C|p.Val540Val|1864/2281|1620/1683|540/560||,G|synonymous_variant|LOW|LOC109102258|LOC109102258|transcript|XM_042771433.1|protein_coding|13/13|c.1620G>C|p.Val540Val|1845/2262|1620/1683|540/560||,G|synonymous_variant|LOW|LOC109102258|LOC109102258|transcript|XM_042771494.1|protein_coding|13/13|c.1614G>C|p.Val538Val|1862/2279|1614/1677|538/558|| | GT:DP:AD:RO:QR:AO:QA:GL | 1/1:2:0,2:0:0:2:74:-7.02588,-0.60206,0 |
| NC_056572.1 | 26171 | . | C | A | 43.0472 | PASS | AB=0.5;ABP=3.0103;AC=1;AF=0.2;AN=2;AO=4;CIGAR=1X;DP=12;DPB=12;DPRA=3;EPP=3.0103;EPPR=3.0103;GTI=0;LEN=1;MEANALT=1;MQM=60;MQMR=60;NS=5;NUMALT=1;ODDS=2.66081;PAIRED=1;PAIREDR=1;PAO=0;PQA=0;PQR=0;PRO=0;QA=148;QR=272;RO=8;RPL=2;RPP=3.0103;RPPR=4.09604;RPR=2;RUN=1;SAF=2;SAP=3.0103;SAR=2;SRF=5;SRP=4.09604;SRR=3;TYPE=snp;technology.illumina=1;ANN=A|intron_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771354.1|protein_coding|11/12|c.1504-390G>T||||||,A|intron_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771433.1|protein_coding|11/12|c.1504-390G>T||||||,A|intron_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771494.1|protein_coding|11/12|c.1504-396G>T|||||| | GT:DP:AD:RO:QR:AO:QA:GL | 0/1:5:2,3:2:50:3:111:-8.84877,0,-3.24459 |
| NC_056572.1 | 26177 | . | A | T | 115.674 | PASS | AB=0.5;ABP=3.0103;AC=1;AF=0.6;AN=2;AO=7;CIGAR=1X;DP=12;DPB=12;DPRA=2.75;EPP=3.32051;EPPR=3.44459;GTI=1;LEN=1;MEANALT=1;MQM=60;MQMR=60;NS=5;NUMALT=1;ODDS=1.20171;PAIRED=1;PAIREDR=1;PAO=0;PQA=0;PQR=0;PRO=0;QA=240;QR=185;RO=5;RPL=4;RPP=3.32051;RPPR=3.44459;RPR=3;RUN=1;SAF=4;SAP=3.32051;SAR=3;SRF=3;SRP=3.44459;SRR=2;TYPE=snp;technology.illumina=1;ANN=T|intron_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771354.1|protein_coding|11/12|c.1504-396T>A||||||,T|intron_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771433.1|protein_coding|11/12|c.1504-396T>A||||||,T|intron_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771494.1|protein_coding|11/12|c.1504-402T>A|||||| | GT:DP:AD:RO:QR:AO:QA:GL | 0/1:5:3,2:3:111:2:67:-4.85738,0,-8.84877 |
| NC_056572.1 | 26849 | . | G | A | 166.204 | PASS | AB=0.5;ABP=3.0103;AC=2;AF=0.7;AN=2;AO=7;CIGAR=1X;DP=9;DPB=9;DPRA=2;EPP=10.7656;EPPR=7.35324;GTI=1;LEN=1;MEANALT=1;MQM=60;MQMR=60;NS=5;NUMALT=1;ODDS=2.51969;PAIRED=1;PAIREDR=1;PAO=0;PQA=0;PQR=0;PRO=0;QA=259;QR=74;RO=2;RPL=5;RPP=5.80219;RPPR=7.35324;RPR=2;RUN=1;SAF=4;SAP=3.32051;SAR=3;SRF=2;SRP=7.35324;SRR=0;TYPE=snp;technology.illumina=1;ANN=A|intron_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771354.1|protein_coding|11/12|c.1503+845C>T||||||,A|intron_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771433.1|protein_coding|11/12|c.1503+845C>T||||||,A|intron_variant|MODIFIER|LOC109102258|LOC109102258|transcript|XM_042771494.1|protein_coding|11/12|c.1503+845C>T|||||| | GT:DP:AD:RO:QR:AO:QA:GL | 1/1:1:0,1:0:0:1:37:-3.69783,-0.30103,0 |
表3.3.1 SNP注释后变异信息记录文件示例
- #CHROM: 变异所在的染色体或者参考序列的名称。
- POS: 变异在染色体上的碱基位置。
- ID: 变异ID。通常为dbSNP数据库中的rs编号,如果没有则为“.”。
- REF: 参考碱基。该位点在参考基因组上的碱基序列。
- ALT: 变异碱基。样本中的碱基序列,如与参考基因组一致则显示为“.”。
- QUAL: 质量得分。变异检测的质量分数。
- FILTER: 过滤状态。表示变异是否通过了质控。
- INFO: 变异额外信息。其中ANN存储了注释信息
- FORMAT: 变异存储格式。定义样本列中数据的顺序。
- Sample: 样本名。
以下为注释信息统计。表格右上角的检索框可用于筛选包含检索内容的条目,即仅显示包含输入关键词的行
| Type | Sample1 | Sample2 | Sample3 | Sample4 | Sample5 |
|---|---|---|---|---|---|
| 3 Prime UTR Variant | 80577 | 80410 | 77360 | 75896 | 80792 |
| 5 Prime UTR Premature Start Codon Gain Variant | 4709 | 4747 | 4643 | 4385 | 4915 |
| 5 Prime UTR Variant | 30352 | 30334 | 29355 | 28327 | 30767 |
| Downstream Gene Variant | 465168 | 465014 | 448146 | 433506 | 466503 |
| Initiator Codon Variant | 6 | 5 | 3 | 5 | 6 |
| Intergenic Region | 494927 | 493813 | 480364 | 461420 | 497948 |
| Intragenic Variant | 41624 | 41637 | 40082 | 38165 | 41904 |
| Intron Variant | 2121594 | 2116136 | 2051847 | 1979818 | 2136267 |
| Missense Variant | 63432 | 63348 | 61637 | 60223 | 65140 |
| Non Coding Transcript Exon Variant | 13312 | 13303 | 12854 | 12492 | 13586 |
| Splice Acceptor Variant | 148 | 168 | 165 | 157 | 163 |
| Splice Donor Variant | 180 | 189 | 167 | 168 | 168 |
| Splice Region Variant | 20590 | 20891 | 19711 | 19352 | 20951 |
| Start Lost | 47 | 53 | 45 | 45 | 47 |
| Stop Gained | 298 | 334 | 306 | 279 | 309 |
| Stop Lost | 73 | 76 | 64 | 74 | 67 |
| Stop Retained Variant | 78 | 67 | 76 | 70 | 57 |
| Synonymous Variant | 120153 | 120582 | 118050 | 114483 | 122475 |
| Upstream Gene Variant | 445054 | 444732 | 426581 | 413668 | 446266 |
表3.3.2 SNP变异信息统计(按对基因的影响预测统计)
- Type: 变异产生了哪些影响,具体释义请参照如下页面的functional-class一节。https://pcingola.github.io/SnpEff/snpeff/inputoutput
| Type | Sample1 | Sample2 | Sample3 | Sample4 | Sample5 |
|---|---|---|---|---|---|
| Downstream | 465168 | 465014 | 448146 | 433506 | 466503 |
| Exon | 194695 | 195044 | 190388 | 184998 | 198846 |
| Intergenic | 494927 | 493813 | 480364 | 461420 | 497948 |
| Intron | 2105174 | 2099401 | 2036142 | 1964465 | 2119646 |
| Splice Site Acceptor | 148 | 168 | 165 | 157 | 163 |
| Splice Site Donor | 178 | 187 | 165 | 166 | 166 |
| Splice Site Region | 19334 | 19619 | 18506 | 18179 | 19654 |
| Transcript | 41624 | 41637 | 40082 | 38165 | 41904 |
| Upstream | 445054 | 444732 | 426581 | 413668 | 446266 |
| UTR 3 Prime | 80577 | 80410 | 77360 | 75896 | 80792 |
| UTR 5 Prime | 35061 | 35081 | 33998 | 32712 | 35682 |
表3.3.3 SNP变异信息统计(按区域统计)
- Type: 变异发生在哪些区域,具体释义请参照如下页面的Variant annotaiton details一节。https://pcingola.github.io/SnpEff/snpeff/inputoutput
| sample | A:T > G:C | A:T > C:G | A:T > T:A | C:G > A:T | C:G > G:C | C:G > T:A |
|---|---|---|---|---|---|---|
| Sample1 | 358452 | 147956 | 204979 | 166569 | 112003 | 454580 |
| Sample2 | 357717 | 147897 | 204876 | 165806 | 111738 | 454079 |
| Sample3 | 351007 | 144524 | 197524 | 159952 | 108062 | 437238 |
| Sample4 | 340870 | 140228 | 191638 | 153023 | 104025 | 419518 |
| Sample5 | 360572 | 148642 | 205503 | 167473 | 112720 | 456976 |
表3.3.4 SNP碱基变化统计
- 本表格展示每个位点碱基变化的统计。如A:T > G:C,表示位点碱基从A:T对变化为G:C对
- 图3.3.1与3.3.2 展示了每个样本中SNP变异的预测饼图, 其中3.3.1是按照变异对基因的影响进行统计,3.3.2是按照变异位点所在区域进行统计。 由于参考基因组注释文件中对单个基因会标记多个transcript(转录本), 所以预测的影响会比变异记录文件具有更多条目数。
- 图3.3.3 展示了SNP变异的碱基变化情况。 其中,X轴表示每个样本,Y轴表示碱基变异情况在样本中出现的次数。 拖拽X轴上的滑条可以自行选择展示范围。
- 图3.3.4 展示了每个样本的SNP变异的密度分布情况。 X轴表示参考基因组中contig的区块,单位为1mb个碱基,Y轴表示每个contig。 变异密度即为每个区块中变异位点的数量除以区块长度,变异密度越高,展示颜色越红。 由于参考基因组中可能具有较多的contig,此处只展示长度排前25的contig。
3.4 INDEL检测与注释
流程结果
├── 04.INDEL (进入文件目录)
│ ├── Sample1
│ │ ├── 表3.4.5_INDEL密度统计_Sample1.csv (因数据过多,未列入报告)
│ │ ├── 图3.4.1_样本INDEL效应类型占比_Sample1.png
│ │ ├── 图3.4.2_样本INDEL发生位点占比_Sample1.png
│ │ └── 图3.4.4_样本INDEL密度热力图_Sample1.png
│ ├── ...
│ ├── 表3.4.2_INDEL变异信息统计(按对基因的影响预测统计).csv
│ ├── 表3.4.3_INDEL变异信息统计(按区域统计).csv
│ ├── 表3.4.4_INDEL长度统计.csv
│ └── 图3.4.3_样本INDEL长度分布.png
└── ...
INDEL变异指的是插入/缺失变异(Insertion and Deletion variation), 即基因组上由一个或多个核苷酸的插入或缺失造成的遗传变异。 由于Freebayes所使用的算法在复杂区域也具有较高的灵敏性与容错率, 所以其在检测SNP位点的同时也可以检测出小片段(不超过50bp)的插入/缺失变异。
INDEL分析中,Freebayes使用的参数与SNP检测相同,过滤参数相同。 下面将针对这一算法在识别SNP与INDEL的步骤差异进行解释:
- 寻找候选变异时,若该区域存在大量错配或低质量比对时,Freebayes会将其标记为INDEL候选位点。
- 变异校验时,由于INDEL涉及连续的碱基变异会使得计算概率空间变得更加复杂, 所以Freebayes会对INDEL候选变异进行局部重比对,以更准确地确定INDEL的位置和类型。 同时,Freebayes会向候选位点左右两侧延伸比对的长度,以确定INDEL的精确边界。
- 输出结果时:由于INDEL可能导致测序读取在不同位置产生相似的比对, Freebayes会进行多重比对以评估最有可能产生的情况。
以下为结果文件示例
| #CHROM | POS | ID | REF | ALT | QUAL | FILTER | INFO | FORMAT | Sample1 |
|---|---|---|---|---|---|---|---|---|---|
| NC_056572.1 | 44914 | . | TTTTTTTTTAAAG | TTTAATTTTTTTTAAAG | 45.0234 | PASS | AB=0.5;ABP=3.0103;AC=1;AF=0.5;AN=2;AO=4;CIGAR=1M4I12M;DP=11;DPB=12.2308;DPRA=0.555556;EPP=5.18177;EPPR=16.0391;GTI=2;LEN=4;MEANALT=1;MQM=60;MQMR=60;NS=5;NUMALT=1;ODDS=0.642093;PAIRED=1;PAIREDR=1;PAO=0;PQA=0;PQR=0;PRO=0;QA=148;QR=210;RO=6;RPL=2;RPP=3.0103;RPPR=4.45795;RPR=2;RUN=1;SAF=1;SAP=5.18177;SAR=3;SRF=2;SRP=4.45795;SRR=4;TYPE=ins;technology.illumina=1;ANN=TTTAATTTTTTTTAAAG|intron_variant|MODIFIER|LOC109080128|LOC109080128|transcript|XM_042771598.1|protein_coding|1/6|c.-10-457_-10-456insAATT|||||| | GT:DP:AD:RO:QR:AO:QA:GL | 0/1:2:1,1:1:37:1:37:-3.09577,0,-3.09577 |
| NC_056572.1 | 51978 | . | CATA | CA | 40.6154 | PASS | AB=0.6;ABP=3.44459;AC=1;AF=0.4;AN=2;AO=4;CIGAR=1M2D1M;DP=10;DPB=8;DPRA=1;EPP=5.18177;EPPR=16.0391;GTI=1;LEN=2;MEANALT=1;MQM=60;MQMR=60;NS=5;NUMALT=1;ODDS=1.33524;PAIRED=1;PAIREDR=1;PAO=0;PQA=0;PQR=0;PRO=0;QA=148;QR=219;RO=6;RPL=2;RPP=3.0103;RPPR=3.0103;RPR=2;RUN=1;SAF=3;SAP=5.18177;SAR=1;SRF=3;SRP=3.0103;SRR=3;TYPE=del;technology.illumina=1;ANN=CA|upstream_gene_variant|MODIFIER|LOC109080128|LOC109080128|transcript|XM_042771598.1|protein_coding||c.-1832_-1831delTA|||||1629|,CA|intergenic_region|MODIFIER|LOC109080128-LOC109080221|LOC109080128-LOC109080221|intergenic_region|LOC109080128-LOC109080221|||n.51980_51981delTA|||||| | GT:DP:AD:RO:QR:AO:QA:GL | 0/1:2:1,1:1:37:1:37:-3.09577,0,-3.09577 |
| NC_056572.1 | 71432 | . | CGA | CTTGA | 79.1264 | PASS | AB=0.5;ABP=3.0103;AC=2;AF=0.5;AN=2;AO=5;CIGAR=1M2I2M;DP=9;DPB=12.3333;DPRA=1.33333;EPP=3.44459;EPPR=5.18177;GTI=2;LEN=2;MEANALT=1;MQM=60;MQMR=60;NS=5;NUMALT=1;ODDS=0.487942;PAIRED=1;PAIREDR=1;PAO=0;PQA=0;PQR=0;PRO=0;QA=185;QR=148;RO=4;RPL=2;RPP=3.44459;RPPR=3.0103;RPR=3;RUN=1;SAF=2;SAP=3.44459;SAR=3;SRF=3;SRP=5.18177;SRR=1;TYPE=ins;technology.illumina=1;ANN=CTTGA|downstream_gene_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771675.1|protein_coding||c.*4039_*4040insAA|||||2644|,CTTGA|downstream_gene_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771824.1|protein_coding||c.*4039_*4040insAA|||||2644|,CTTGA|downstream_gene_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771758.1|protein_coding||c.*4039_*4040insAA|||||2644|,CTTGA|downstream_gene_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771715.1|protein_coding||c.*4039_*4040insAA|||||2644|,CTTGA|intergenic_region|MODIFIER|LOC109080128-LOC109080221|LOC109080128-LOC109080221|intergenic_region|LOC109080128-LOC109080221|||n.71432_71433insTT|||||| | GT:DP:AD:RO:QR:AO:QA:GL | 1/1:1:0,1:0:0:1:37:-3.69783,-0.30103,0 |
| NC_056572.1 | 90469 | . | TAACAAAAG | TAG | 37.6244 | PASS | AB=0.5;ABP=3.0103;AC=1;AF=0.4;AN=2;AO=4;CIGAR=1M6D2M;DP=10;DPB=7.33333;DPRA=1.55556;EPP=5.18177;EPPR=3.0103;GTI=1;LEN=6;MEANALT=1;MQM=60;MQMR=60;NS=5;NUMALT=1;ODDS=1.33524;PAIRED=1;PAIREDR=1;PAO=0;PQA=0;PQR=0;PRO=0;QA=148;QR=209;RO=6;RPL=2;RPP=3.0103;RPPR=4.45795;RPR=2;RUN=1;SAF=3;SAP=5.18177;SAR=1;SRF=3;SRP=3.0103;SRR=3;TYPE=del;technology.illumina=1;ANN=TAG|intron_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771675.1|protein_coding|17/23|c.2560+267_2560+272delTTTTGT||||||,TAG|intron_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771824.1|protein_coding|15/21|c.2083+267_2083+272delTTTTGT||||||,TAG|intron_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771758.1|protein_coding|15/21|c.2128+267_2128+272delTTTTGT||||||,TAG|intron_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771715.1|protein_coding|15/21|c.2341+267_2341+272delTTTTGT|||||| | GT:DP:AD:RO:QR:AO:QA:GL | 0/1:4:2,2:2:74:2:74:-5.82176,0,-5.82176 |
| NC_056572.1 | 118123 | . | CAG | CAAG | 151.211 | PASS | AB=0.428571;ABP=3.32051;AC=2;AF=0.5;AN=2;AO=8;CIGAR=1M1I2M;DP=15;DPB=17.6667;DPRA=2.66667;EPP=4.09604;EPPR=3.32051;GTI=2;LEN=1;MEANALT=1;MQM=60;MQMR=60;NS=5;NUMALT=1;ODDS=0.0873481;PAIRED=1;PAIREDR=1;PAO=0;PQA=0;PQR=0;PRO=0;QA=284;QR=259;RO=7;RPL=4;RPP=3.0103;RPPR=5.80219;RPR=4;RUN=1;SAF=5;SAP=4.09604;SAR=3;SRF=4;SRP=3.32051;SRR=3;TYPE=ins;technology.illumina=1;ANN=CAAG|upstream_gene_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771824.1|protein_coding||c.-1106_-1105insT|||||576|,CAAG|intron_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771675.1|protein_coding|3/23|c.508-7831_508-7830insT||||||,CAAG|intron_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771758.1|protein_coding|1/21|c.75+2741_75+2742insT||||||,CAAG|intron_variant|MODIFIER|LOC109080221|LOC109080221|transcript|XM_042771715.1|protein_coding|3/21|c.508-7831_508-7830insT|||||| | GT:DP:AD:RO:QR:AO:QA:GL | 1/1:3:0,3:0:0:3:111:-10.3539,-0.90309,0 |
表3.4.1 INDEL注释后变异信息记录文件示例
- #CHROM: 变异所在的染色体或者参考序列的名称。
- POS: 变异在染色体上的碱基位置。
- ID: 变异ID。通常为dbSNP数据库中的rs编号,如果没有则为“.”。
- REF: 参考基因组序列。该位点在参考基因组上的碱基序列。
- ALT: 变异序列。样本中的碱基序列,如与参考基因组一致则显示为“.”。
- QUAL: 质量得分。变异检测的质量分数。
- FILTER: 过滤状态。表示变异是否通过了质控。
- INFO: 变异额外信息。其中ANN存储了注释信息
- FORMAT: 变异存储格式。定义样本列中数据的顺序。
- Sample: 样本名。
以下为注释信息统计。表格右上角的检索框可用于筛选包含检索内容的条目,即仅显示包含输入关键词的行
| Type | Sample1 | Sample2 | Sample3 | Sample4 | Sample5 |
|---|---|---|---|---|---|
| 3 Prime UTR Variant | 9459 | 9508 | 8903 | 8827 | 9546 |
| 5 Prime UTR Truncation | 1 | 0 | 1 | 0 | 1 |
| 5 Prime UTR Variant | 2734 | 2776 | 2648 | 2527 | 2735 |
| Conservative Inframe Deletion | 308 | 327 | 306 | 307 | 308 |
| Conservative Inframe Insertion | 310 | 371 | 355 | 353 | 373 |
| Disruptive Inframe Deletion | 234 | 236 | 219 | 215 | 240 |
| Disruptive Inframe Insertion | 253 | 242 | 209 | 212 | 234 |
| Downstream Gene Variant | 44603 | 45286 | 42998 | 41789 | 44964 |
| Exon Loss Variant | 1 | 0 | 1 | 0 | 1 |
| Frameshift Variant | 483 | 478 | 421 | 410 | 479 |
| Intergenic Region | 42405 | 42594 | 41271 | 39965 | 42729 |
| Intragenic Variant | 3433 | 3493 | 3270 | 3129 | 3475 |
| Intron Variant | 193018 | 193809 | 185206 | 177681 | 192779 |
| Non Coding Transcript Exon Variant | 901 | 943 | 888 | 851 | 925 |
| Non Coding Transcript Variant | 26 | 32 | 31 | 20 | 33 |
| Splice Acceptor Variant | 127 | 136 | 119 | 118 | 134 |
| Splice Donor Variant | 98 | 104 | 82 | 104 | 108 |
| Splice Region Variant | 1591 | 1647 | 1635 | 1496 | 1660 |
| Start Lost | 10 | 12 | 13 | 12 | 9 |
| Start Retained Variant | 2 | 1 | 2 | 2 | 2 |
| Stop Gained | 11 | 11 | 12 | 6 | 12 |
| Stop Lost | 3 | 3 | 3 | 3 | 3 |
| Stop Retained Variant | 12 | 13 | 11 | 12 | 11 |
| Upstream Gene Variant | 42128 | 42454 | 40167 | 39504 | 42236 |
表3.4.2 INDEL变异信息统计(按对基因的影响预测统计)
- Type: 变异产生了哪些影响,具体释义请参照如下页面的functional-class一节。https://pcingola.github.io/SnpEff/snpeff/inputoutput
| Type | Sample1 | Sample2 | Sample3 | Sample4 | Sample5 |
|---|---|---|---|---|---|
| Downstream | 44603 | 45286 | 42998 | 41789 | 44964 |
| Exon | 2480 | 2590 | 2391 | 2339 | 2550 |
| Intergenic | 42405 | 42594 | 41271 | 39965 | 42729 |
| Intron | 191380 | 192121 | 183546 | 176139 | 191067 |
| Splice Site Acceptor | 123 | 132 | 116 | 117 | 130 |
| Splice Site Donor | 92 | 99 | 76 | 99 | 102 |
| Splice Site Region | 1504 | 1553 | 1554 | 1413 | 1561 |
| Transcript | 3459 | 3525 | 3301 | 3149 | 3508 |
| Upstream | 42128 | 42454 | 40167 | 39504 | 42236 |
| UTR 3 Prime | 9459 | 9508 | 8903 | 8827 | 9546 |
| UTR 5 Prime | 2734 | 2775 | 2648 | 2526 | 2735 |
表3.4.3 INDEL变异信息统计(按区域统计)
- Type: 变异发生在哪些区域,具体释义请参照如下页面的Variant annotaiton details一节。https://pcingola.github.io/SnpEff/snpeff/inputoutput
- 图3.4.1与3.4.2 展示了每个样本中INDEL变异的预测饼图, 其中3.4.1是按照变异对基因的影响进行统计,3.4.2是按照变异位点所在区域进行统计。 由于参考基因组注释文件中对单个基因会标记多个transcript(转录本), 所以预测的影响会比变异记录文件具有更多条目数。
- 图3.4.3 展示了每个样本中INDEL变异的长度分布情况。 其中X轴展示了INDEL长度,Y轴展示了变异数量。不同的样本以不同颜色的折线表示。
- 图3.3.4 展示了每个样本的SNP变异的密度分布情况。 X轴表示参考基因组中contig的区块,单位为1mb个碱基,Y轴表示每个contig。 变异密度即为每个区块中变异位点的数量除以区块长度,变异密度越高,展示颜色越红。 由于参考基因组中可能具有较多的contig,此处只展示长度排前25的contig。
3.5 SV检测与注释
流程结果
├── 05.SV (进入文件目录)
│ ├── Sample1
│ │ ├── 表3.5.5_SV长度统计_Sample1.csv (因数据过多,未列入报告)
│ │ ├── 图3.5.1_样本SV效应类型占比_Sample1.png
│ │ └── 图3.5.2_样本SV发生位点占比_Sample1.png
│ ├── ...
│ ├── 表3.5.2_SV变异信息统计(按对基因的影响预测统计).csv
│ ├── 表3.5.3_SV变异信息统计(按区域统计).csv
│ ├── 表3.5.4_SV变异类型统计.csv
│ ├── 图3.5.3_样本SV类型堆叠图.png
│ └── 图3.5.4_样本SV长度分布箱线图.png
└── ...
SV(Structural Variants)指的是染色体结构上变异,包括插入(Insertion)、删除(Deletion)、倒位(Inversion)、易位(Translocation)等。它们在基因组中占据较大的区域,与一些疾病的发生、遗传差异及进化等都有关联。 由于算法限制,对SNP进行检测的算法对50bp以上的插入缺失事件的检测精度不高,且无法检测倒位等大规模事件。 为了更全面地检测变异,本次分析采用Delly软件[5]对所有样本的比对数据(bam)进行结构变异检测。 检测到的SV结果经过过滤后进行注释,以确保结果的准确性和可靠性。
Delly的算法与参数介绍如下:
- Delly是一种配对末端测序数据(Paired-End reads)和分割序列(Split reads)的结构变异检测软件。分割序列指的是序列中有部分碱基没有连续比对至参考基因组上的序列,这通常是由某种结构性变异产生。 在这种情况下,比对算法会将这些没能比对的碱基标记为软裁剪(soft-clipped)以确保一段序列中的多数碱基被成功比对。通过结合前者所提供的序列方向与预期距离,与后者提供的断点(Breakend)位置, Delly可以精确定位结构性变异的发生位置,从而提供更全面的变异信息。
- 该算法的计算步骤如下:
- 提取异常配对信息:根据测序序列对的预期距离(来源于制备时文库片段长度)和预期方向性,识别比对至参考基因组后发生异常的读取对。如序列对分别比对至不同的染色体上,这也被视作异常。
- 定义潜在结构变异区域:基于每一种结构性变异在比对时具有的特性定义结构性变异的潜在发生区域。
- 分割序列定位:在定义的潜在区域中寻找分割序列,并利用分割序列两端的断点初步定位结构变异的发生位置。
- 局部重比对: 基于分割序列产生k-mer短序列,再与参考基因组进行局部重比对。局部重比对后,根据证据reads建立共识序列(consensus sequence)。 共识序列在此处指的是由重合区域出现频率最高的碱基连接而成的一短序列。
- 最终比对: 使用动态双重规划,将共识序列与潜在结构变异区域进行对齐,并在正反链方向上计算评分矩阵以定义最优左右断点。
- 结果输出:将SV输出至vcf文件,其中每个变异的质量评分和其它相关信息都会被记录。
- Delly的运行参数解释如下:
- map-qual: 参与SV检测的reads所应具有的最低比对质量。(默认为1,当前为1)
- qual-tra: 作为移位证据的reads所应具有的最低质量分数(默认为20,当前为20)
- mad-cutoff: 过滤插入大小异常的reads时采取的最大中位绝对偏差(MAD)(默认为9,当前为9)
- minclip: 计入split-read时,reads中的soft-clipped长度下限(默认为25,当前为25)
- min-clique-size: 成为SV支持证据的最小reads配对数量(默认为2,当前为2)
- minrefsep: split-reads在参考基因组上的最小分离距离(默认为25,当前为25)。
- maxreadsep: 计入同一个SV的两个split-reads之间的最大分离距离(默认为40,当前为40)。
在检测完成后,需要对变异位点进行过滤,以保证结果的可信度。本次分析中变异位点的过滤规则如下:
- 保留QUAL分数大于20的位点(推荐值为:30)
- 保留PE > 3的位点(推荐值为:3)
- PE表示为SV预测提供支持的配对末端(Paired Ends)数量
- 保留SR > 0的位点(推荐值为:0)
- SR表示为SV预测提供支持的分割序列(Split reads)数量
- 保留SRQ > 0的位点(推荐值为:0)
- SR表示为SV预测提供支持的分割序列(Split reads)的比对质量
以下为结果文件示例
| #CHROM | POS | ID | REF | ALT | QUAL | FILTER | INFO | FORMAT | Sample1 |
|---|---|---|---|---|---|---|---|---|---|
| NC_056572.1 | 4054080 | DEL00000095 | T | <DEL> | 420.0 | PASS | PRECISE;SVTYPE=DEL;SVMETHOD=EMBL.DELLYv1.1.6;END=4054596;PE=4;MAPQ=60;CT=3to5;CIPOS=-3,3;CIEND=-3,3;SRMAPQ=60;INSLEN=0;HOMLEN=2;SR=3;SRQ=0.965986;CONSENSUS=AACTCTTATTAAATTTAGTCTGTGGTCCTTTACATACACTAGTGCTTTAATTAGAATTAGTGCAGAATCTGGGGCAGCAGAATGTAAAATTTAGTTTTGTCAAATATTCCATTTTAATTATACTTCTGTATATTCAAATTGCAATC;CE=1.85884;AC=2;AN=2;ANN=<DEL>|intergenic_region|MODIFIER|LOC109096534-frg1|LOC109096534-frg1|intergenic_region|LOC109096534-frg1|||n.4054081_4054596del|||||| | GT:GL:GQ:FT:RCL:RC:RCR:RDCN:DR:DV:RR:RV | 1/1:-17.9995,-1.5046,0:15:PASS:489:297:1039:0:0:4:0:5 |
| NC_056572.1 | 5093081 | DEL00000122 | T | <DEL> | 420.0 | PASS | PRECISE;SVTYPE=DEL;SVMETHOD=EMBL.DELLYv1.1.6;END=5096112;PE=3;MAPQ=60;CT=3to5;CIPOS=-5,5;CIEND=-5,5;SRMAPQ=60;INSLEN=0;HOMLEN=4;SR=4;SRQ=0.987805;CONSENSUS=TACCAGTGACATTTAAAATGAGGAAAAATAACTAGCCCAAACATGTTGGAATTTTTAATCTTGAAAAATACAAGCACAATTTCTTAAGATTGCATAATATATAAATTAATAATACTTTCCAGAGCCTTTATGGCCTTGAGTACATACCAGCTGAAATATGGAAT;CE=1.86206;AC=2;AN=2;ANN=<DEL>|intergenic_region|MODIFIER|LOC109078388-LOC109090214|LOC109078388-LOC109090214|intergenic_region|LOC109078388-LOC109090214|||n.5093082_5096112del|||||| | GT:GL:GQ:FT:RCL:RC:RCR:RDCN:DR:DV:RR:RV | 1/1:-13.9994,-1.20351,0:12:LowQual:7:0:9:0:0:3:0:4 |
| NC_056572.1 | 5288221 | DEL00000134 | A | <DEL> | 360.0 | PASS | PRECISE;SVTYPE=DEL;SVMETHOD=EMBL.DELLYv1.1.6;END=13586398;PE=3;MAPQ=60;CT=3to5;CIPOS=-1,1;CIEND=-1,1;SRMAPQ=60;INSLEN=0;HOMLEN=0;SR=3;SRQ=0.972603;CONSENSUS=AAGAATGCATGCATTTGCACAAGAGGTAGCTTGACCCCAATCCACAGCTGTTTTTGTCATAGCTATAATAATGCAAATTAAAATGGTGCAGTTTCCTATTAACTGGCTCTGTCCAATTAATAGGTCACTGTGCATCTGTCAGCAGT;CE=1.96876;AC=2;AN=2;ANN=<DEL>|chromosome_number_variation|HIGH|||chromosome|NC_056572.1|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109095492|LOC109095492|gene_variant|LOC109095492|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC122134481|LOC122134481|gene_variant|LOC122134481|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109079371|LOC109079371|gene_variant|LOC109079371|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109095406|LOC109095406|gene_variant|LOC109095406|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109095415|LOC109095415|gene_variant|LOC109095415|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109091060|LOC109091060|gene_variant|LOC109091060|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109084847|LOC109084847|gene_variant|LOC109084847|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122137050|LOC122137050|gene_variant|LOC122137050|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109068102|LOC109068102|gene_variant|LOC109068102|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109078080|LOC109078080|gene_variant|LOC109078080|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122146593|LOC122146593|gene_variant|LOC122146593|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109078329|LOC109078329|gene_variant|LOC109078329|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109078237|LOC109078237|gene_variant|LOC109078237|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109100816|LOC109100816|gene_variant|LOC109100816|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109052457|LOC109052457|gene_variant|LOC109052457|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109051718|LOC109051718|gene_variant|LOC109051718|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109051716|LOC109051716|gene_variant|LOC109051716|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109079158|LOC109079158|gene_variant|LOC109079158|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109079183|LOC109079183|gene_variant|LOC109079183|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109113423|LOC109113423|gene_variant|LOC109113423|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC122146595|LOC122146595|gene_variant|LOC122146595|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109083632|LOC109083632|gene_variant|LOC109083632|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109086113|LOC109086113|gene_variant|LOC109086113|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109090665|LOC109090665|gene_variant|LOC109090665|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109094653|LOC109094653|gene_variant|LOC109094653|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109090659|LOC109090659|gene_variant|LOC109090659|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109090651|LOC109090651|gene_variant|LOC109090651|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109072040|LOC109072040|gene_variant|LOC109072040|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109109310|LOC109109310|gene_variant|LOC109109310|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109053664|LOC109053664|gene_variant|LOC109053664|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109108964|LOC109108964|gene_variant|LOC109108964|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109108968|LOC109108968|gene_variant|LOC109108968|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109108976|LOC109108976|gene_variant|LOC109108976|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109108963|LOC109108963|gene_variant|LOC109108963|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109108965|LOC109108965|gene_variant|LOC109108965|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|xpa|xpa|gene_variant|xpa|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109108971|LOC109108971|gene_variant|LOC109108971|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC122137963|LOC122137963|gene_variant|LOC122137963|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109108981|LOC109108981|gene_variant|LOC109108981|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|slc24a2|slc24a2|gene_variant|slc24a2|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|tspan5b|tspan5b|gene_variant|tspan5b|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|grk4|grk4|gene_variant|grk4|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|snx8b|snx8b|gene_variant|snx8b|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|iqce|iqce|gene_variant|iqce|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109069582|LOC109069582|gene_variant|LOC109069582|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109069581|LOC109069581|gene_variant|LOC109069581|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109069577|LOC109069577|gene_variant|LOC109069577|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|si:dkey-26i13.8|si:dkey-26i13.8|gene_variant|si:dkey-26i13.8|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109070447|LOC109070447|gene_variant|LOC109070447|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109082845|LOC109082845|gene_variant|LOC109082845|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109072834|LOC109072834|gene_variant|LOC109072834|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109084254|LOC109084254|gene_variant|LOC109084254|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109061405|LOC109061405|gene_variant|LOC109061405|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC122146727|LOC122146727|gene_variant|LOC122146727|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109069038|LOC109069038|gene_variant|LOC109069038|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109082538|LOC109082538|gene_variant|LOC109082538|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109081867|LOC109081867|gene_variant|LOC109081867|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109081865|LOC109081865|gene_variant|LOC109081865|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109100245|LOC109100245|gene_variant|LOC109100245|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109086090|LOC109086090|gene_variant|LOC109086090|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109086096|LOC109086096|gene_variant|LOC109086096|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109086091|LOC109086091|gene_variant|LOC109086091|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109086079|LOC109086079|gene_variant|LOC109086079|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109081871|LOC109081871|gene_variant|LOC109081871|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109066378|LOC109066378|gene_variant|LOC109066378|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109066379|LOC109066379|gene_variant|LOC109066379|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109088128|LOC109088128|gene_variant|LOC109088128|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109066750|LOC109066750|gene_variant|LOC109066750|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109063029|LOC109063029|gene_variant|LOC109063029|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109064567|LOC109064567|gene_variant|LOC109064567|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109088132|LOC109088132|gene_variant|LOC109088132|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109077144|LOC109077144|gene_variant|LOC109077144|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109088130|LOC109088130|gene_variant|LOC109088130|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109088135|LOC109088135|gene_variant|LOC109088135|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109111928|LOC109111928|gene_variant|LOC109111928|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122138707|LOC122138707|gene_variant|LOC122138707|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109085059|LOC109085059|gene_variant|LOC109085059|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109085056|LOC109085056|gene_variant|LOC109085056|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109085058|LOC109085058|gene_variant|LOC109085058|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109085054|LOC109085054|gene_variant|LOC109085054|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109090858|LOC109090858|gene_variant|LOC109090858|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122138786|LOC122138786|gene_variant|LOC122138786|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122138813|LOC122138813|gene_variant|LOC122138813|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122138844|LOC122138844|gene_variant|LOC122138844|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109074881|LOC109074881|gene_variant|LOC109074881|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122145905|LOC122145905|gene_variant|LOC122145905|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109074880|LOC109074880|gene_variant|LOC109074880|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC122146597|LOC122146597|gene_variant|LOC122146597|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109074885|LOC109074885|gene_variant|LOC109074885|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109074882|LOC109074882|gene_variant|LOC109074882|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109083105|LOC109083105|gene_variant|LOC109083105|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109083127|LOC109083127|gene_variant|LOC109083127|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109063886|LOC109063886|gene_variant|LOC109063886|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109077732|LOC109077732|gene_variant|LOC109077732|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109100636|LOC109100636|gene_variant|LOC109100636|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109064065|LOC109064065|gene_variant|LOC109064065|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109094368|LOC109094368|gene_variant|LOC109094368|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109063976|LOC109063976|gene_variant|LOC109063976|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109068501|LOC109068501|gene_variant|LOC109068501|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109094284|LOC109094284|gene_variant|LOC109094284|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109068012|LOC109068012|gene_variant|LOC109068012|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109062389|LOC109062389|gene_variant|LOC109062389|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109065094|LOC109065094|gene_variant|LOC109065094|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC122139250|LOC122139250|gene_variant|LOC122139250|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109095481|LOC109095481|gene_variant|LOC109095481|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109095401|LOC109095401|gene_variant|LOC109095401|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109093640|LOC109093640|gene_variant|LOC109093640|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109060541|LOC109060541|gene_variant|LOC109060541|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109060387|LOC109060387|gene_variant|LOC109060387|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109052115|LOC109052115|gene_variant|LOC109052115|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109108622|LOC109108622|gene_variant|LOC109108622|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109051882|LOC109051882|gene_variant|LOC109051882|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109051902|LOC109051902|gene_variant|LOC109051902|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109051883|LOC109051883|gene_variant|LOC109051883|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109051881|LOC109051881|gene_variant|LOC109051881|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109086892|LOC109086892|gene_variant|LOC109086892|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109086894|LOC109086894|gene_variant|LOC109086894|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109086893|LOC109086893|gene_variant|LOC109086893|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC122139615|LOC122139615|gene_variant|LOC122139615|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109068184|LOC109068184|gene_variant|LOC109068184|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109068183|LOC109068183|gene_variant|LOC109068183|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109068182|LOC109068182|gene_variant|LOC109068182|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109068180|LOC109068180|gene_variant|LOC109068180|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122145967|LOC122145967|gene_variant|LOC122145967|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109068179|LOC109068179|gene_variant|LOC109068179|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122146383|LOC122146383|gene_variant|LOC122146383|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109053958|LOC109053958|gene_variant|LOC109053958|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109052588|LOC109052588|gene_variant|LOC109052588|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109052585|LOC109052585|gene_variant|LOC109052585|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109096812|LOC109096812|gene_variant|LOC109096812|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109069351|LOC109069351|gene_variant|LOC109069351|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109095694|LOC109095694|gene_variant|LOC109095694|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109095059|LOC109095059|gene_variant|LOC109095059|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109110991|LOC109110991|gene_variant|LOC109110991|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC122146818|LOC122146818|gene_variant|LOC122146818|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109098228|LOC109098228|gene_variant|LOC109098228|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109055793|LOC109055793|gene_variant|LOC109055793|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109071097|LOC109071097|gene_variant|LOC109071097|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109071105|LOC109071105|gene_variant|LOC109071105|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109064231|LOC109064231|gene_variant|LOC109064231|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109064193|LOC109064193|gene_variant|LOC109064193|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109064236|LOC109064236|gene_variant|LOC109064236|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109064239|LOC109064239|gene_variant|LOC109064239|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109064184|LOC109064184|gene_variant|LOC109064184|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122146602|LOC122146602|gene_variant|LOC122146602|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109052551|LOC109052551|gene_variant|LOC109052551|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109097173|LOC109097173|gene_variant|LOC109097173|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122146506|LOC122146506|gene_variant|LOC122146506|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122140031|LOC122140031|gene_variant|LOC122140031|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109055833|LOC109055833|gene_variant|LOC109055833|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109077898|LOC109077898|gene_variant|LOC109077898|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109090362|LOC109090362|gene_variant|LOC109090362|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109045087|LOC109045087|gene_variant|LOC109045087|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109044867|LOC109044867|gene_variant|LOC109044867|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109057260|LOC109057260|gene_variant|LOC109057260|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109057259|LOC109057259|gene_variant|LOC109057259|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109092346|LOC109092346|gene_variant|LOC109092346|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109086150|LOC109086150|gene_variant|LOC109086150|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109060930|LOC109060930|gene_variant|LOC109060930|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109049467|LOC109049467|gene_variant|LOC109049467|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC122146783|LOC122146783|gene_variant|LOC122146783|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109098823|LOC109098823|gene_variant|LOC109098823|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109089105|LOC109089105|gene_variant|LOC109089105|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109098990|LOC109098990|gene_variant|LOC109098990|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC122140343|LOC122140343|gene_variant|LOC122140343|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109099299|LOC109099299|gene_variant|LOC109099299|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109074794|LOC109074794|gene_variant|LOC109074794|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109066109|LOC109066109|gene_variant|LOC109066109|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|ndfip2|ndfip2|gene_variant|ndfip2|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109074737|LOC109074737|gene_variant|LOC109074737|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122140423|LOC122140423|gene_variant|LOC122140423|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109070584|LOC109070584|gene_variant|LOC109070584|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109068129|LOC109068129|gene_variant|LOC109068129|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109061870|LOC109061870|gene_variant|LOC109061870|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122147300|LOC122147300|gene_variant|LOC122147300|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC122147450|LOC122147450|gene_variant|LOC122147450|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109054415|LOC109054415|gene_variant|LOC109054415|||n.13586398_5288222del||||||,<DEL>|feature_ablation|HIGH|LOC109062975|LOC109062975|gene_variant|LOC109062975|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109112091|LOC109112091|gene_variant|LOC109112091|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109065683|LOC109065683|gene_variant|LOC109065683|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC122147574|LOC122147574|gene_variant|LOC122147574|||n.5288222_13586398del||||||,<DEL>|feature_ablation|HIGH|LOC109044867&LOC109045087&LOC109049467&LOC109051716&LOC109051718&LOC109051881&LOC109051882&LOC109051883&LOC109051902&LOC109052115&LOC109052457&LOC109052551&LOC109052585&LOC109052588&LOC109053664&LOC109053958&LOC109054415&LOC109055793&LOC109055833&LOC109057259&LOC109057260&LOC109060387&LOC109060541&LOC109060930&LOC109061405&LOC109061870&LOC109062389&LOC109062975&LOC109063029&LOC109063886&LOC109063976&LOC109064065&LOC109064184&LOC109064193&LOC109064231&LOC109064236&LOC109064239&LOC109064567&LOC109065094&LOC109065683&LOC109066109&LOC109066378&LOC109066379&LOC109066750&LOC109068012&LOC109068102&LOC109068129&LOC109068179&LOC109068180&LOC109068182&LOC109068183&LOC109068184&LOC109068501&LOC109069038&LOC109069351&LOC109069577&LOC109069581&LOC109069582&LOC109070447&LOC109070584&LOC109071097&LOC109071105&LOC109072040&LOC109072834&LOC109074737&LOC109074794&LOC109074880&LOC109074881&LOC109074882&LOC109074885&LOC109077144&LOC109077732&LOC109077898&LOC109078080&LOC109078237&LOC109078329&LOC109079158&LOC109079183&LOC109079371&LOC109081865&LOC109081867&LOC109081871&LOC109082538&LOC109082845&LOC109083105&LOC109083127&LOC109083632&LOC109084254&LOC109084847&LOC109085054&LOC109085056&LOC109085058&LOC109085059&LOC109086079&LOC109086090&LOC109086091&LOC109086096&LOC109086113&LOC109086150&LOC109086892&LOC109086893&LOC109086894&LOC109088128&LOC109088130&LOC109088132&LOC109088135&LOC109089105&LOC109090362&LOC109090651&LOC109090659&LOC109090665&LOC109090858&LOC109091060&LOC109092346&LOC109093640&LOC109094284&LOC109094368&LOC109094653&LOC109095059&LOC109095401&LOC109095406&LOC109095415&LOC109095481&LOC109095492&LOC109095694&LOC109096812&LOC109097173&LOC109098228&LOC109098823&LOC109098990&LOC109099299&LOC109100245&LOC109100636&LOC109100816&LOC109108622&LOC109108963&LOC109108964&LOC109108965&LOC109108968&LOC109108971&LOC109108976&LOC109108981&LOC109109310&LOC109110991&LOC109111928&LOC109112091&LOC109113423&LOC122134481&LOC122137050&LOC122137963&LOC122138707&LOC122138786&LOC122138813&LOC122138844&LOC122139250&LOC122139615&LOC122140031&LOC122140343&LOC122140423&LOC122145905&LOC122145967&LOC122146383&LOC122146506&LOC122146593&LOC122146595&LOC122146597&LOC122146602&LOC122146727&LOC122146783&LOC122146818&LOC122147300&LOC122147450&LOC122147574&grk4&iqce&ndfip2&si:dkey-26i13.8&slc24a2&snx8b&tspan5b&xpa|LOC109044867&LOC109045087&LOC109049467&LOC109051716&LOC109051718&LOC109051881&LOC109051882&LOC109051883&LOC109051902&LOC109052115&LOC109052457&LOC109052551&LOC109052585&LOC109052588&LOC109053664&LOC109053958&LOC109054415&LOC109055793&LOC109055833&LOC109057259&LOC109057260&LOC109060387&LOC109060541&LOC109060930&LOC109061405&LOC109061870&LOC109062389&LOC109062975&LOC109063029&LOC109063886&LOC109063976&LOC109064065&LOC109064184&LOC109064193&LOC109064231&LOC109064236&LOC109064239&LOC109064567&LOC109065094&LOC109065683&LOC109066109&LOC109066378&LOC109066379&LOC109066750&LOC109068012&LOC109068102&LOC109068129&LOC109068179&LOC109068180&LOC109068182&LOC109068183&LOC109068184&LOC109068501&LOC109069038&LOC109069351&LOC109069577&LOC109069581&LOC109069582&LOC109070447&LOC109070584&LOC109071097&LOC109071105&LOC109072040&LOC109072834&LOC109074737&LOC109074794&LOC109074880&LOC109074881&LOC109074882&LOC109074885&LOC109077144&LOC109077732&LOC109077898&LOC109078080&LOC109078237&LOC109078329&LOC109079158&LOC109079183&LOC109079371&LOC109081865&LOC109081867&LOC109081871&LOC109082538&LOC109082845&LOC109083105&LOC109083127&LOC109083632&LOC109084254&LOC109084847&LOC109085054&LOC109085056&LOC109085058&LOC109085059&LOC109086079&LOC109086090&LOC109086091&LOC109086096&LOC109086113&LOC109086150&LOC109086892&LOC109086893&LOC109086894&LOC109088128&LOC109088130&LOC109088132&LOC109088135&LOC109089105&LOC109090362&LOC109090651&LOC109090659&LOC109090665&LOC109090858&LOC109091060&LOC109092346&LOC109093640&LOC109094284&LOC109094368&LOC109094653&LOC109095059&LOC109095401&LOC109095406&LOC109095415&LOC109095481&LOC109095492&LOC109095694&LOC109096812&LOC109097173&LOC109098228&LOC109098823&LOC109098990&LOC109099299&LOC109100245&LOC109100636&LOC109100816&LOC109108622&LOC109108963&LOC109108964&LOC109108965&LOC109108968&LOC109108971&LOC109108976&LOC109108981&LOC109109310&LOC109110991&LOC109111928&LOC109112091&LOC109113423&LOC122134481&LOC122137050&LOC122137963&LOC122138707&LOC122138786&LOC122138813&LOC122138844&LOC122139250&LOC122139615&LOC122140031&LOC122140343&LOC122140423&LOC122145905&LOC122145967&LOC122146383&LOC122146506&LOC122146593&LOC122146595&LOC122146597&LOC122146602&LOC122146727&LOC122146783&LOC122146818&LOC122147300&LOC122147450&LOC122147574&grk4&iqce&ndfip2&si:dkey-26i13.8&slc24a2&snx8b&tspan5b&xpa|||||||||||,<DEL>|transcript_ablation|HIGH|LOC109095492|LOC109095492|transcript|XM_042721120.1|protein_coding|2/7|c.-2508_*8286767del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122134481|LOC122134481|transcript|XM_042721422.1|protein_coding|4/4|c.-12882_*8280166del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109079371|LOC109079371|transcript|XM_042721374.1|protein_coding|1/16|c.-8266496_*22048del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109095406|LOC109095406|transcript|XM_019109117.2|protein_coding|10/11|c.-8254851_*38298del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109095415|LOC109095415|transcript|XM_019109127.2|protein_coding|2/6|c.-8244093_*51308del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109091060|LOC109091060|transcript|XM_042721978.1|protein_coding|5/17|c.-8231986_*55206del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109084847|LOC109084847|transcript|XM_042758691.1|protein_coding|1/1|c.-8226469_*70526del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068102|LOC109068102|transcript|XM_042722478.1|protein_coding|8/14|c.-8206510_*81338del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109078080|LOC109078080|transcript|XM_042722659.1|protein_coding|2/5|c.-8188516_*104716del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122146593|LOC122146593|transcript|XM_042766228.1|protein_coding|2/2|c.-8187219_*110237del|p.0?|||||WARNING_TRANSCRIPT_NO_START_CODON,<DEL>|transcript_ablation|HIGH|LOC109078237|LOC109078237|transcript|XM_019093573.2|protein_coding|2/4|c.-8163232_*134164del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100816|LOC109100816|transcript|XM_042722938.1|protein_coding|1/22|c.-142261_*8121526del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109052457|LOC109052457|transcript|XM_042766235.1|protein_coding|17/46|c.-8052474_*180112del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109051718|LOC109051718|transcript|XM_042723338.1|protein_coding|8/8|c.-8046218_*247657del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109051716|LOC109051716|transcript|XM_019069200.2|protein_coding|2/5|c.-252820_*8040625del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109079158|LOC109079158|transcript|XM_042723676.1|protein_coding|9/9|c.-261086_*8016602del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109079183|LOC109079183|transcript|XM_019094354.2|protein_coding|2/3|c.-8005793_*284590del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109113423|LOC109113423|transcript|XM_042724364.1|protein_coding|4/4|c.-605627_*7685479del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109083632|LOC109083632|transcript|XM_042724937.1|protein_coding|1/4|c.-7323980_*960750del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086113|LOC109086113|transcript|XM_042725707.1|protein_coding|2/5|c.-7157932_*1138614del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109090665|LOC109090665|transcript|XM_042725191.1|protein_coding|21/22|c.-1140663_*7144024del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109094653|LOC109094653|transcript|XM_042725475.1|protein_coding|1/7|c.-7134868_*1155692del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109090659|LOC109090659|transcript|XM_042725372.1|protein_coding|3/6|c.-7130370_*1164092del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109090651|LOC109090651|transcript|XM_042725869.1|protein_coding|11/13|c.-7114440_*1169162del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109072040|LOC109072040|transcript|XM_042726278.1|protein_coding|2/4|c.-1199847_*7091379del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109109310|LOC109109310|transcript|XM_042726771.1|protein_coding|3/11|c.-7079478_*1210372del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109053664|LOC109053664|transcript|XM_042726710.1|protein_coding|2/18|c.-7069427_*1219678del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109108964|LOC109108964|transcript|XM_042726944.1|protein_coding|15/18|c.-7060668_*1229976del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109108968|LOC109108968|transcript|XM_019122085.2|protein_coding|6/6|c.-1241392_*7048422del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109108976|LOC109108976|transcript|XM_019122093.2|protein_coding|1/4|c.-7044854_*1252664del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109108963|LOC109108963|transcript|XM_042727131.1|protein_coding|2/18|c.-1257061_*7029709del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109108965|LOC109108965|transcript|XM_019122081.2|protein_coding|1/2|c.-7018557_*1272803del|p.0?|||||,<DEL>|transcript_ablation|HIGH|xpa|xpa|transcript|XM_042727341.1|protein_coding|4/7|c.-7009570_*1284961del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109108971|LOC109108971|transcript|XM_019122088.2|protein_coding|3/4|c.-1290735_*7006390del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122137963|LOC122137963|transcript|XR_006155179.1|pseudogene|2/2|n.-1292588_*7004313del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109108981|LOC109108981|transcript|XM_042727545.1|protein_coding|4/6|c.-7000476_*1293944del|p.0?|||||,<DEL>|transcript_ablation|HIGH|slc24a2|slc24a2|transcript|XM_042727802.1|protein_coding|1/10|c.-6983089_*1301332del|p.0?|||||,<DEL>|transcript_ablation|HIGH|tspan5b|tspan5b|transcript|XM_042727990.1|protein_coding|5/9|c.-6968818_*1322004del|p.0?|||||,<DEL>|transcript_ablation|HIGH|grk4|grk4|transcript|XM_042728095.1|protein_coding|17/17|c.-1332684_*6943591del|p.0?|||||,<DEL>|transcript_ablation|HIGH|snx8b|snx8b|transcript|XM_042760598.1|protein_coding|11/12|c.-6933013_*1360042del|p.0?|||||,<DEL>|transcript_ablation|HIGH|iqce|iqce|transcript|XM_042728383.1|protein_coding|17/21|c.-1367342_*6922958del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109069582|LOC109069582|transcript|XM_019086209.2|protein_coding|2/15|c.-1377123_*6894465del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109069581|LOC109069581|transcript|XM_019086208.2|protein_coding|2/2|c.-1409991_*6886278del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109069577|LOC109069577|transcript|XM_042766261.1|protein_coding|1/1|c.-6872884_*1424762del|p.0?|||||,<DEL>|transcript_ablation|HIGH|si:dkey-26i13.8|si:dkey-26i13.8|transcript|XM_042728781.1|protein_coding|11/21|c.-6854872_*1427543del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109070447|LOC109070447|transcript|XM_042728945.1|protein_coding|10/18|c.-6845980_*1445163del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109082845|LOC109082845|transcript|XM_042728876.1|protein_coding|1/15|c.-1452603_*6837168del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109072834|LOC109072834|transcript|XM_042729031.1|protein_coding|1/4|c.-6825736_*1462127del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109084254|LOC109084254|transcript|XM_042729213.1|protein_coding|2/2|c.-1483829_*6811769del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109061405|LOC109061405|transcript|XM_042729635.1|protein_coding|21/47|c.-1536753_*6556612del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122146727|LOC122146727|transcript|XR_006161248.1|snRNA|1/1|n.-1621425_*6676589del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109069038|LOC109069038|transcript|XR_006155431.1|pseudogene|3/3|n.-1665118_*6632026del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109082538|LOC109082538|transcript|XM_042729451.1|protein_coding|2/2|c.-1685529_*6608070del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109081867|LOC109081867|transcript|XM_042729953.1|protein_coding|8/8|c.-6549756_*1746205del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109081865|LOC109081865|transcript|XM_042729799.1|protein_coding|20/26|c.-1748888_*6536434del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100245|LOC109100245|transcript|XM_042730632.1|protein_coding|72/79|c.-1767771_*6430404del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086090|LOC109086090|transcript|XM_042730723.1|protein_coding|1/7|c.-1871273_*6422730del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086096|LOC109086096|transcript|XM_042766272.1|protein_coding|7/22|c.-1876938_*6413461del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086091|LOC109086091|transcript|XM_042730835.1|protein_coding|1/7|c.-6404349_*1888750del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086079|LOC109086079|transcript|XM_042730916.1|protein_coding|3/6|c.-6385104_*1908267del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109081871|LOC109081871|transcript|XM_019096835.2|protein_coding|5/5|c.-6361954_*1931200del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109066378|LOC109066378|transcript|XM_042731024.1|protein_coding|3/5|c.-1936593_*6353010del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109066379|LOC109066379|transcript|XM_042731402.1|protein_coding|2/5|c.-1950486_*6345302del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109088128|LOC109088128|transcript|XM_042731529.1|protein_coding|4/11|c.-1964477_*6259256del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109066750|LOC109066750|transcript|XM_042731632.1|protein_coding|6/6|c.-2043596_*6251825del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109063029|LOC109063029|transcript|XM_042731753.1|protein_coding|11/12|c.-6211939_*2052198del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064567|LOC109064567|transcript|XM_042731966.1|protein_coding|9/12|c.-2107893_*6175881del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109088132|LOC109088132|transcript|XM_019092874.2|protein_coding|3/7|c.-6166838_*2125737del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109077144|LOC109077144|transcript|XM_042732023.1|protein_coding|2/9|c.-6160700_*2133286del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109088130|LOC109088130|transcript|XM_042732107.1|protein_coding|1/8|c.-6152974_*2137813del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109088135|LOC109088135|transcript|XM_019092625.2|protein_coding|5/5|c.-6129215_*2165548del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109111928|LOC109111928|transcript|XM_019124929.2|protein_coding|1/6|c.-6092125_*2199628del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122138707|LOC122138707|transcript|XM_042732615.1|protein_coding|1/1|c.-6087896_*2209765del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109085059|LOC109085059|transcript|XM_042732732.1|protein_coding|2/2|c.-6083964_*2212509del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109085056|LOC109085056|transcript|XM_042732837.1|protein_coding|15/20|c.-6066954_*2216623del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109085058|LOC109085058|transcript|XM_019099696.2|protein_coding|9/9|c.-2234971_*6055372del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109085054|LOC109085054|transcript|XM_042733096.1|protein_coding|12/12|c.-6041610_*2246002del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109090858|LOC109090858|transcript|XM_019104697.2|protein_coding|1/6|c.-6037676_*2258733del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122138786|LOC122138786|transcript|XM_042733400.1|protein_coding|2/4|c.-5979220_*2313201del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122138813|LOC122138813|transcript|XM_042733449.1|protein_coding|6/6|c.-5946719_*2349786del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122138844|LOC122138844|transcript|XM_042733538.1|protein_coding|3/5|c.-5896318_*2395388del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109074881|LOC109074881|transcript|XM_042733648.1|protein_coding|2/11|c.-5890267_*2403595del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122145905|LOC122145905|transcript|XM_042762219.1|protein_coding|2/2|c.-5887301_*2409440del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109074880|LOC109074880|transcript|XM_019090888.2|protein_coding|3/13|c.-2413515_*5874711del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122146597|LOC122146597|transcript|XM_042766297.1|protein_coding|2/9|c.-5865344_*2423750del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109074885|LOC109074885|transcript|XM_042734203.1|protein_coding|1/8|c.-2433773_*5859708del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109074882|LOC109074882|transcript|XM_019090891.2|protein_coding|2/8|c.-5855251_*2439800del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109083105|LOC109083105|transcript|XM_042733836.1|protein_coding|7/18|c.-2443385_*5786681del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109083127|LOC109083127|transcript|XM_042734511.1|protein_coding|1/3|c.-5691919_*2526733del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109063886|LOC109063886|transcript|XM_042734671.1|protein_coding|2/2|c.-2556391_*5737379del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109077732|LOC109077732|transcript|XM_042734708.1|protein_coding|16/20|c.-2619964_*5661552del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100636|LOC109100636|transcript|XM_042734768.1|protein_coding|2/17|c.-2642066_*5642858del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064065|LOC109064065|transcript|XM_042734876.1|protein_coding|3/3|c.-5554682_*2739928del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109094368|LOC109094368|transcript|XM_042735040.1|protein_coding|2/18|c.-2752299_*5536084del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109063976|LOC109063976|transcript|XM_042735345.1|protein_coding|2/11|c.-2764011_*5530435del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068501|LOC109068501|transcript|XM_042735545.1|protein_coding|2/3|c.-2771884_*5525655del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109094284|LOC109094284|transcript|XM_042735430.1|protein_coding|20/22|c.-5512319_*2772916del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068012|LOC109068012|transcript|XM_042735646.1|protein_coding|2/22|c.-2787685_*5497950del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109062389|LOC109062389|transcript|XM_042735774.1|protein_coding|8/11|c.-5489497_*2803490del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122139250|LOC122139250|transcript|XM_042735871.1|protein_coding|4/4|c.-2820633_*5475954del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109095481|LOC109095481|transcript|XM_042735958.1|protein_coding|10/11|c.-2825159_*5464614del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109095401|LOC109095401|transcript|XM_042766319.1|protein_coding|3/5|c.-5459069_*2834273del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109093640|LOC109093640|transcript|XM_042736040.1|protein_coding|17/17|c.-5415700_*2846789del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109060541|LOC109060541|transcript|XM_042736164.1|protein_coding|1/2|c.-5405696_*2889228del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109060387|LOC109060387|transcript|XM_019077585.2|protein_coding|1/2|c.-2898102_*5397980del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109052115|LOC109052115|transcript|XM_042736590.1|protein_coding|2/14|c.-2921480_*5286649del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109108622|LOC109108622|transcript|XM_042736760.1|protein_coding|3/9|c.-2933178_*5354685del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109051882|LOC109051882|transcript|XM_019069376.2|protein_coding|4/4|c.-5253441_*3037620del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109051902|LOC109051902|transcript|XM_019069389.2|protein_coding|1/5|c.-5239711_*3053457del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109051883|LOC109051883|transcript|XR_006156346.1|pseudogene|4/4|n.-5232609_*3060536del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109051881|LOC109051881|transcript|XM_019069375.2|protein_coding|6/6|c.-3065750_*5229437del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086892|LOC109086892|transcript|XM_042737688.1|protein_coding|4/5|c.-3071562_*5222373del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086894|LOC109086894|transcript|XM_042737816.1|protein_coding|2/11|c.-5209155_*3080409del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086893|LOC109086893|transcript|XM_042737908.1|protein_coding|19/21|c.-3098269_*5189506del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122139615|LOC122139615|transcript|XM_042738061.1|protein_coding|1/1|c.-5178604_*3119000del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068184|LOC109068184|transcript|XM_042738157.1|protein_coding|5/5|c.-5170755_*3122570del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068183|LOC109068183|transcript|XM_019085034.2|protein_coding|1/4|c.-5165049_*3130005del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068182|LOC109068182|transcript|XM_019085033.2|protein_coding|1/4|c.-3136932_*5157205del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068180|LOC109068180|transcript|XM_042766334.1|protein_coding|26/33|c.-5106797_*3147752del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122145967|LOC122145967|transcript|XR_006160735.1|pseudogene|1/2|n.-5058697_*3239042del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068179|LOC109068179|transcript|XM_042763822.1|protein_coding|2/5|c.-5052332_*3243477del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122146383|LOC122146383|transcript|XM_042765019.1|protein_coding|2/2|c.-5046894_*3249844del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109053958|LOC109053958|transcript|XM_042738868.1|protein_coding|4/5|c.-3259829_*5026629del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109052588|LOC109052588|transcript|XM_042738664.1|protein_coding|15/15|c.-5001048_*3274314del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109052585|LOC109052585|transcript|XM_042738521.1|protein_coding|2/5|c.-3282078_*5010050del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109096812|LOC109096812|transcript|XM_042738956.1|protein_coding|11/22|c.-3305613_*4944129del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109069351|LOC109069351|transcript|XM_019086081.2|protein_coding|5/7|c.-3357111_*4926546del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109095694|LOC109095694|transcript|XM_042739373.1|protein_coding|7/7|c.-3375591_*4910891del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109095059|LOC109095059|transcript|XM_042739467.1|protein_coding|1/7|c.-3394043_*4887574del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109110991|LOC109110991|transcript|XM_042739567.1|protein_coding|8/10|c.-3414323_*4876109del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122146818|LOC122146818|transcript|XR_006161277.1|snoRNA|1/1|n.-3413770_*4884323del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109098228|LOC109098228|transcript|XM_042739881.1|protein_coding|6/17|c.-3428519_*4851733del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109055793|LOC109055793|transcript|XM_019072980.2|protein_coding|1/2|c.-4770877_*3523817del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109071097|LOC109071097|transcript|XM_019087568.2|protein_coding|4/4|c.-3534583_*4756029del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109071105|LOC109071105|transcript|XM_042740255.1|protein_coding|4/5|c.-3544887_*4744763del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064231|LOC109064231|transcript|XM_019081222.2|protein_coding|7/12|c.-3578087_*4713297del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064193|LOC109064193|transcript|XM_042740705.1|protein_coding|1/6|c.-4709930_*3586481del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064236|LOC109064236|transcript|XM_042740983.1|protein_coding|2/6|c.-4704787_*3589981del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064239|LOC109064239|transcript|XM_042741081.1|protein_coding|11/11|c.-4684221_*3604106del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064184|LOC109064184|transcript|XM_042741326.1|protein_coding|22/27|c.-4457904_*3657713del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122146602|LOC122146602|transcript|XM_042766348.1|protein_coding|1/5|c.-3870522_*4398871del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109052551|LOC109052551|transcript|XM_042741243.1|protein_coding|19/27|c.-4077588_*4043389del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109097173|LOC109097173|transcript|XM_042741523.1|protein_coding|1/10|c.-4062196_*4224782del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122146506|LOC122146506|transcript|XR_006161080.1|pseudogene|1/3|n.-4053439_*4239754del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122140031|LOC122140031|transcript|XR_006156760.1|pseudogene|1/3|n.-4008996_*4286167del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109055833|LOC109055833|transcript|XR_006156764.1|pseudogene|5/5|n.-4293664_*4002237del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109077898|LOC109077898|transcript|XM_019093218.2|protein_coding|1/4|c.-4337120_*3925752del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109090362|LOC109090362|transcript|XM_042741885.1|protein_coding|8/29|c.-3875982_*4391692del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109044867|LOC109044867|transcript|XM_042741960.1|protein_coding|10/11|c.-3842201_*4450732del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109057260|LOC109057260|transcript|XM_042742332.1|protein_coding|1/5|c.-4457636_*3838838del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109057259|LOC109057259|transcript|XM_042742230.1|protein_coding|3/9|c.-3820410_*4464154del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086150|LOC109086150|transcript|XM_042742547.1|protein_coding|4/20|c.-3792577_*4493826del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109060930|LOC109060930|transcript|XM_042742648.1|protein_coding|1/23|c.-4540523_*3588843del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109049467|LOC109049467|transcript|XM_042743037.1|protein_coding|3/17|c.-4736331_*3495463del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109098823|LOC109098823|transcript|XM_042766667.1|protein_coding|1/9|c.-3480390_*4812867del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109089105|LOC109089105|transcript|XR_006156958.1|pseudogene|4/4|n.-3468675_*4826705del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109098990|LOC109098990|transcript|XM_042743267.1|protein_coding|28/46|c.-4841175_*3432774del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122140343|LOC122140343|transcript|XM_042743557.1|protein_coding|1/15|c.-3422833_*4866814del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109099299|LOC109099299|transcript|XM_042767389.1|protein_coding|3/3|c.-4893842_*3400259del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109074794|LOC109074794|transcript|XM_042743791.1|protein_coding|4/8|c.-3380380_*4911936del|p.0?|||||WARNING_TRANSCRIPT_MULTIPLE_STOP_CODONS,<DEL>|transcript_ablation|HIGH|LOC109066109|LOC109066109|transcript|XM_042743640.1|protein_coding|1/4|c.-3367652_*4918843del|p.0?|||||,<DEL>|transcript_ablation|HIGH|ndfip2|ndfip2|transcript|XM_042766368.1|protein_coding|11/11|c.-4934870_*3338270del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109074737|LOC109074737|transcript|XM_019090756.2|protein_coding|1/2|c.-3299804_*4995924del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122140423|LOC122140423|transcript|XR_006157103.1|pseudogene|1/2|n.-2455512_*5841087del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109070584|LOC109070584|transcript|XM_042744082.1|protein_coding|1/9|c.-5903044_*2391479del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109061870|LOC109061870|transcript|XM_019078948.2|protein_coding|1/1|c.-1998466_*6297623del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122147300|LOC122147300|transcript|XR_006161604.1|pseudogene|3/4|n.-1882190_*6410356del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122147450|LOC122147450|transcript|XR_006161658.1|pseudogene|4/4|n.-6499476_*1794552del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109054415|LOC109054415|transcript|XM_042744269.1|protein_coding|2/2|c.-1694093_*6588329del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109062975|LOC109062975|transcript|XM_019080033.2|protein_coding|1/2|c.-6753946_*1540232del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109112091|LOC109112091|transcript|XM_042744486.1|protein_coding|2/9|c.-7467585_*624876del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122147574|LOC122147574|transcript|XR_006161732.1|pseudogene|3/3|n.-7975734_*322110del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109095492|LOC109095492|transcript|XM_042721264.1|protein_coding|2/6|c.-2508_*8286767del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109095492|LOC109095492|transcript|XM_042721192.1|protein_coding|2/7|c.-2508_*8286767del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109095492|LOC109095492|transcript|XM_042721298.1|protein_coding|2/6|c.-2508_*8286767del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122134481|LOC122134481|transcript|XM_042721626.1|protein_coding|4/4|c.-12879_*8280166del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122134481|LOC122134481|transcript|XM_042721501.1|protein_coding|4/5|c.-12930_*8280166del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122134481|LOC122134481|transcript|XM_042721566.1|protein_coding|4/5|c.-12927_*8280166del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122134481|LOC122134481|transcript|XM_042721449.1|protein_coding|4/4|c.-13090_*8280166del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122134481|LOC122134481|transcript|XM_042721484.1|protein_coding|4/4|c.-13087_*8280166del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109095406|LOC109095406|transcript|XM_042721789.1|protein_coding|10/11|c.-8254976_*38298del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109091060|LOC109091060|transcript|XM_042722056.1|protein_coding|5/17|c.-8231986_*55206del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109091060|LOC109091060|transcript|XM_042722137.1|protein_coding|5/17|c.-8231986_*55206del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109091060|LOC109091060|transcript|XM_042722210.1|protein_coding|5/16|c.-8231986_*55206del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109091060|LOC109091060|transcript|XM_042722275.1|protein_coding|5/16|c.-8231986_*55206del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068102|LOC109068102|transcript|XM_042722582.1|protein_coding|8/14|c.-8208538_*81338del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068102|LOC109068102|transcript|XM_042722521.1|protein_coding|8/15|c.-8206836_*81338del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100816|LOC109100816|transcript|XM_042722974.1|protein_coding|1/21|c.-142261_*8122356del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100816|LOC109100816|transcript|XM_042723014.1|protein_coding|1/22|c.-142261_*8121526del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100816|LOC109100816|transcript|XM_042723044.1|protein_coding|1/21|c.-142261_*8121526del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100816|LOC109100816|transcript|XM_042723088.1|protein_coding|1/21|c.-142261_*8121526del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100816|LOC109100816|transcript|XM_042723226.1|protein_coding|1/20|c.-142261_*8121526del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100816|LOC109100816|transcript|XM_042723151.1|protein_coding|1/20|c.-142261_*8121526del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109051718|LOC109051718|transcript|XM_042723392.1|protein_coding|8/8|c.-8046868_*247657del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109051716|LOC109051716|transcript|XM_042723491.1|protein_coding|2/4|c.-252820_*8040625del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109051716|LOC109051716|transcript|XM_042723568.1|protein_coding|2/5|c.-254938_*8040625del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109079158|LOC109079158|transcript|XM_042723818.1|protein_coding|9/10|c.-261189_*8016602del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109079158|LOC109079158|transcript|XM_042723892.1|protein_coding|9/10|c.-261156_*8016602del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109079158|LOC109079158|transcript|XM_042723963.1|protein_coding|9/10|c.-261166_*8016602del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109079158|LOC109079158|transcript|XM_042724095.1|protein_coding|9/9|c.-262885_*8016602del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109079158|LOC109079158|transcript|XM_042723745.1|protein_coding|9/9|c.-267132_*8016602del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109079158|LOC109079158|transcript|XM_042724035.1|protein_coding|9/9|c.-267614_*8016602del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109079158|LOC109079158|transcript|XM_019094323.2|protein_coding|7/7|c.-275307_*8016602del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109113423|LOC109113423|transcript|XM_042724431.1|protein_coding|4/4|c.-605627_*7685479del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109113423|LOC109113423|transcript|XM_042724574.1|protein_coding|4/4|c.-605627_*7685479del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109113423|LOC109113423|transcript|XM_042724507.1|protein_coding|4/4|c.-605627_*7685479del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109083632|LOC109083632|transcript|XM_042725071.1|protein_coding|1/6|c.-7323980_*959004del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109083632|LOC109083632|transcript|XM_042725135.1|protein_coding|1/6|c.-7323980_*958880del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109083632|LOC109083632|transcript|XM_042724992.1|protein_coding|1/5|c.-7323980_*959142del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086113|LOC109086113|transcript|XM_019100605.2|protein_coding|2/6|c.-7157600_*1138614del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109090665|LOC109090665|transcript|XM_042725225.1|protein_coding|21/22|c.-1140832_*7144024del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109090665|LOC109090665|transcript|XM_042725283.1|protein_coding|21/22|c.-1141249_*7144024del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109094653|LOC109094653|transcript|XM_019108382.2|protein_coding|1/6|c.-7134868_*1155692del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109094653|LOC109094653|transcript|XM_019108390.2|protein_coding|1/5|c.-7134868_*1155692del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109090651|LOC109090651|transcript|XM_042726038.1|protein_coding|7/12|c.-7114440_*1169162del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109090651|LOC109090651|transcript|XM_042726113.1|protein_coding|7/12|c.-7104843_*1169162del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109090651|LOC109090651|transcript|XM_042726176.1|protein_coding|11/13|c.-7104867_*1169162del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109090651|LOC109090651|transcript|XM_042725954.1|protein_coding|11/13|c.-7100401_*1169162del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109072040|LOC109072040|transcript|XM_042726341.1|protein_coding|2/4|c.-1198292_*7091379del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109072040|LOC109072040|transcript|XM_042726614.1|protein_coding|2/4|c.-1198580_*7091379del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109072040|LOC109072040|transcript|XM_042726485.1|protein_coding|2/4|c.-1199962_*7091379del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109072040|LOC109072040|transcript|XM_042726414.1|protein_coding|2/4|c.-1200136_*7091379del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109072040|LOC109072040|transcript|XM_042726540.1|protein_coding|2/4|c.-1200378_*7091379del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109109310|LOC109109310|transcript|XM_042726874.1|protein_coding|3/10|c.-7079283_*1210332del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109109310|LOC109109310|transcript|XM_042726817.1|protein_coding|3/11|c.-7079283_*1210372del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109109310|LOC109109310|transcript|XM_042726852.1|protein_coding|3/10|c.-7079478_*1210332del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109108981|LOC109108981|transcript|XM_042727607.1|protein_coding|4/6|c.-7000476_*1293944del|p.0?|||||,<DEL>|transcript_ablation|HIGH|slc24a2|slc24a2|transcript|XM_042727877.1|protein_coding|1/9|c.-6983089_*1302039del|p.0?|||||,<DEL>|transcript_ablation|HIGH|grk4|grk4|transcript|XM_042728155.1|protein_coding|17/17|c.-1332684_*6943646del|p.0?|||||,<DEL>|transcript_ablation|HIGH|grk4|grk4|transcript|XM_019091508.2|protein_coding|10/16|c.-1332684_*6943591del|p.0?|||||,<DEL>|transcript_ablation|HIGH|grk4|grk4|transcript|XM_019091556.2|protein_coding|10/16|c.-1332684_*6943646del|p.0?|||||,<DEL>|transcript_ablation|HIGH|snx8b|snx8b|transcript|XM_042761300.1|protein_coding|11/11|c.-6932949_*1360042del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109069582|LOC109069582|transcript|XM_042728555.1|protein_coding|2/14|c.-1377123_*6894465del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109072834|LOC109072834|transcript|XM_042729098.1|protein_coding|1/4|c.-6825736_*1462127del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109072834|LOC109072834|transcript|XM_042729126.1|protein_coding|1/5|c.-6826038_*1462127del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109084254|LOC109084254|transcript|XM_042729342.1|protein_coding|2/2|c.-1482954_*6811769del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109084254|LOC109084254|transcript|XM_042729278.1|protein_coding|2/2|c.-1482987_*6811769del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109084254|LOC109084254|transcript|XM_042729278.1|protein_coding|2/2|c.-1482987_*6811769del|p.0?|||||ERROR_OUT_OF_CHROMOSOME_RANGE,<DEL>|transcript_ablation|HIGH|LOC109082538|LOC109082538|transcript|XM_042729522.1|protein_coding|2/2|c.-1685529_*6608076del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109081867|LOC109081867|transcript|XM_042730096.1|protein_coding|8/8|c.-6550014_*1746205del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109081867|LOC109081867|transcript|XM_042730023.1|protein_coding|8/8|c.-6549751_*1746205del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109081867|LOC109081867|transcript|XM_042730149.1|protein_coding|1/7|c.-6549756_*1746205del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109081865|LOC109081865|transcript|XM_042729862.1|protein_coding|20/26|c.-1748888_*6536434del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100245|LOC109100245|transcript|XM_042730255.1|protein_coding|12/32|c.-1822203_*6430404del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100245|LOC109100245|transcript|XM_042730323.1|protein_coding|12/31|c.-1822203_*6430404del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100245|LOC109100245|transcript|XM_042730386.1|protein_coding|12/33|c.-1822203_*6430505del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100245|LOC109100245|transcript|XM_042730415.1|protein_coding|12/33|c.-1822203_*6430505del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100245|LOC109100245|transcript|XM_042730446.1|protein_coding|12/32|c.-1822203_*6430505del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100245|LOC109100245|transcript|XM_042730531.1|protein_coding|12/31|c.-1822203_*6430547del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100245|LOC109100245|transcript|XR_006155495.1|pseudogene|12/32|n.-1822112_*6430228del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100245|LOC109100245|transcript|XM_042730566.1|protein_coding|12/17|c.-1839428_*6430404del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109081871|LOC109081871|transcript|XM_019096838.2|protein_coding|5/5|c.-6361954_*1931200del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109081871|LOC109081871|transcript|XM_019096837.2|protein_coding|5/6|c.-6361851_*1931200del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109081871|LOC109081871|transcript|XM_042731249.1|protein_coding|5/6|c.-6360220_*1931200del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109063029|LOC109063029|transcript|XM_042731897.1|protein_coding|11/12|c.-6211939_*2052198del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109063029|LOC109063029|transcript|XM_042731827.1|protein_coding|11/12|c.-6211939_*2052198del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109088135|LOC109088135|transcript|XM_042732358.1|protein_coding|5/5|c.-6129215_*2165539del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109111928|LOC109111928|transcript|XM_042732514.1|protein_coding|1/7|c.-6092125_*2199628del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109085056|LOC109085056|transcript|XM_042732906.1|protein_coding|15/21|c.-6066954_*2216487del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109074885|LOC109074885|transcript|XM_042734280.1|protein_coding|1/8|c.-2433776_*5859708del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109083105|LOC109083105|transcript|XM_042734039.1|protein_coding|7/18|c.-2443385_*5816594del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109083105|LOC109083105|transcript|XM_042733980.1|protein_coding|7/18|c.-2443385_*5803232del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109083105|LOC109083105|transcript|XM_042734105.1|protein_coding|7/18|c.-2443385_*5795185del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109083105|LOC109083105|transcript|XM_042733908.1|protein_coding|7/18|c.-2443382_*5786681del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109083127|LOC109083127|transcript|XM_042734572.1|protein_coding|1/4|c.-5692033_*2526733del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109100636|LOC109100636|transcript|XM_042734822.1|protein_coding|2/18|c.-2642066_*5642858del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064065|LOC109064065|transcript|XM_042734932.1|protein_coding|3/3|c.-5554682_*2739928del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109094368|LOC109094368|transcript|XM_042735186.1|protein_coding|2/17|c.-2752299_*5536958del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109094368|LOC109094368|transcript|XM_042735118.1|protein_coding|2/17|c.-2752299_*5536084del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109094368|LOC109094368|transcript|XM_042735256.1|protein_coding|2/18|c.-2752299_*5536084del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109093640|LOC109093640|transcript|XM_042736112.1|protein_coding|17/17|c.-5415700_*2843506del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109060541|LOC109060541|transcript|XM_042736235.1|protein_coding|1/3|c.-5405047_*2889228del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109060387|LOC109060387|transcript|XM_019077651.2|protein_coding|1/2|c.-2896765_*5397980del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109060387|LOC109060387|transcript|XM_042736469.1|protein_coding|1/2|c.-2898075_*5397980del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109052115|LOC109052115|transcript|XM_042736661.1|protein_coding|2/14|c.-2921915_*5286649del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109051902|LOC109051902|transcript|XM_042736953.1|protein_coding|1/6|c.-5239711_*3053425del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109051883|LOC109051883|transcript|XR_002011443.2|pseudogene|4/4|n.-5232609_*3060536del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086892|LOC109086892|transcript|XM_042737726.1|protein_coding|4/5|c.-3071562_*5222373del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109086893|LOC109086893|transcript|XM_042737967.1|protein_coding|19/19|c.-3098269_*5190240del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068182|LOC109068182|transcript|XM_042738428.1|protein_coding|1/4|c.-3136932_*5158118del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109068179|LOC109068179|transcript|XR_006160887.1|pseudogene|2/5|n.-5052257_*3243151del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109052588|LOC109052588|transcript|XM_042738832.1|protein_coding|13/13|c.-5003419_*3274314del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109052588|LOC109052588|transcript|XM_042738735.1|protein_coding|15/15|c.-5001312_*3274314del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109052588|LOC109052588|transcript|XM_042738798.1|protein_coding|15/15|c.-4998321_*3274314del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109052585|LOC109052585|transcript|XR_006156467.1|pseudogene|2/6|n.-3281202_*5010460del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109096812|LOC109096812|transcript|XM_042739016.1|protein_coding|11/22|c.-3305613_*4944129del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109096812|LOC109096812|transcript|XM_042739091.1|protein_coding|11/21|c.-3305613_*4944129del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109096812|LOC109096812|transcript|XM_042739159.1|protein_coding|11/21|c.-3305613_*4944129del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109110991|LOC109110991|transcript|XM_042739787.1|protein_coding|11/11|c.-3414323_*4875942del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109110991|LOC109110991|transcript|XM_042739709.1|protein_coding|8/8|c.-3414939_*4876109del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109110991|LOC109110991|transcript|XM_042739640.1|protein_coding|8/9|c.-3414968_*4876109del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109098228|LOC109098228|transcript|XM_042739947.1|protein_coding|6/16|c.-3428188_*4851733del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109098228|LOC109098228|transcript|XM_042740069.1|protein_coding|6/18|c.-3428519_*4851733del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109098228|LOC109098228|transcript|XM_042740021.1|protein_coding|6/17|c.-3428519_*4851733del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109098228|LOC109098228|transcript|XM_042740118.1|protein_coding|6/16|c.-3428519_*4851733del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109098228|LOC109098228|transcript|XM_042740190.1|protein_coding|6/17|c.-3428519_*4851733del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109055793|LOC109055793|transcript|XM_019072981.2|protein_coding|1/2|c.-4770877_*3523817del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064193|LOC109064193|transcript|XM_042740781.1|protein_coding|1/6|c.-4709930_*3586481del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064193|LOC109064193|transcript|XM_042740890.1|protein_coding|1/5|c.-4709930_*3586481del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064193|LOC109064193|transcript|XM_042740847.1|protein_coding|1/5|c.-4709930_*3586481del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109064239|LOC109064239|transcript|XM_042741144.1|protein_coding|11/11|c.-4684221_*3604119del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109077898|LOC109077898|transcript|XM_042741805.1|protein_coding|1/4|c.-4350928_*3925752del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109044867|LOC109044867|transcript|XM_042742039.1|protein_coding|10/11|c.-3842201_*4450732del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109060930|LOC109060930|transcript|XM_042742791.1|protein_coding|1/23|c.-4540523_*3588843del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109060930|LOC109060930|transcript|XM_042742723.1|protein_coding|1/23|c.-4540523_*3588843del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109060930|LOC109060930|transcript|XM_042742931.1|protein_coding|1/20|c.-4603065_*3588843del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109060930|LOC109060930|transcript|XM_042742868.1|protein_coding|1/20|c.-4609899_*3588843del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109049467|LOC109049467|transcript|XM_042743076.1|protein_coding|3/16|c.-4736331_*3501584del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109049467|LOC109049467|transcript|XM_042743114.1|protein_coding|3/11|c.-4736331_*3514535del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109098990|LOC109098990|transcript|XM_042743340.1|protein_coding|28/45|c.-4841175_*3432774del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109098990|LOC109098990|transcript|XM_042743375.1|protein_coding|28/46|c.-4841175_*3432774del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109098990|LOC109098990|transcript|XM_042743436.1|protein_coding|28/45|c.-4841175_*3432774del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109099299|LOC109099299|transcript|XM_042768028.1|protein_coding|3/3|c.-4893842_*3400259del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109066109|LOC109066109|transcript|XM_042743691.1|protein_coding|1/3|c.-3377207_*4918843del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109074737|LOC109074737|transcript|XM_019090757.2|protein_coding|1/2|c.-3299796_*4995924del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC109112091|LOC109112091|transcript|XM_042744560.1|protein_coding|2/9|c.-7468736_*624876del|p.0?|||||,<DEL>|intragenic_variant|MODIFIER|LOC122137050|LOC122137050|gene_variant|LOC122137050|||n.5288222_13586398del||||||,<DEL>|intragenic_variant|MODIFIER|LOC109078329|LOC109078329|gene_variant|LOC109078329|||n.5288222_13586398del||||||,<DEL>|intragenic_variant|MODIFIER|LOC122146595|LOC122146595|gene_variant|LOC122146595|||n.5288222_13586398del||||||,<DEL>|intragenic_variant|MODIFIER|LOC109065094|LOC109065094|gene_variant|LOC109065094|||n.5288222_13586398del||||||,<DEL>|intragenic_variant|MODIFIER|LOC109045087|LOC109045087|gene_variant|LOC109045087|||n.5288222_13586398del||||||,<DEL>|intragenic_variant|MODIFIER|LOC109092346|LOC109092346|gene_variant|LOC109092346|||n.5288222_13586398del||||||,<DEL>|intragenic_variant|MODIFIER|LOC122146783|LOC122146783|gene_variant|LOC122146783|||n.5288222_13586398del||||||,<DEL>|intragenic_variant|MODIFIER|LOC109068129|LOC109068129|gene_variant|LOC109068129|||n.5288222_13586398del||||||,<DEL>|intragenic_variant|MODIFIER|LOC109065683|LOC109065683|gene_variant|LOC109065683|||n.5288222_13586398del||||||;LOF=(LOC109086090|LOC109086090|1|1.00),(LOC109086079|LOC109086079|1|1.00),(LOC109090858|LOC109090858|1|1.00),(LOC109069577|LOC109069577|1|1.00),(LOC109109310|LOC109109310|4|1.00),(LOC109088135|LOC109088135|2|1.00),(LOC109086894|LOC109086894|1|1.00),(iqce|iqce|1|1.00),(LOC109085059|LOC109085059|1|1.00),(LOC109085058|LOC109085058|1|1.00),(LOC109112091|LOC109112091|2|1.00),(LOC109052457|LOC109052457|1|1.00),(LOC109110991|LOC109110991|4|1.00),(LOC109097173|LOC109097173|1|1.00),(LOC109090665|LOC109090665|3|1.00),(LOC109053958|LOC109053958|1|1.00),(LOC109055793|LOC109055793|2|1.00),(LOC109084254|LOC109084254|3|1.00),(LOC109098228|LOC109098228|6|1.00),(LOC109108965|LOC109108965|1|1.00),(LOC109090651|LOC109090651|5|1.00),(LOC109064065|LOC109064065|2|1.00),(LOC109053664|LOC109053664|1|1.00),(snx8b|snx8b|2|1.00),(LOC122146597|LOC122146597|1|1.00),(LOC109083105|LOC109083105|5|1.00),(LOC109079371|LOC109079371|1|1.00),(LOC109086892|LOC109086892|2|1.00),(LOC109069581|LOC109069581|1|1.00),(LOC109074880|LOC109074880|1|1.00),(LOC109051718|LOC109051718|2|1.00),(LOC109095059|LOC109095059|1|1.00),(LOC109086096|LOC109086096|1|1.00),(LOC109066379|LOC109066379|1|1.00),(LOC122139250|LOC122139250|1|1.00),(LOC109068501|LOC109068501|1|1.00),(LOC109095401|LOC109095401|1|1.00),(LOC109082845|LOC109082845|1|1.00),(LOC109083632|LOC109083632|4|1.00),(LOC109086091|LOC109086091|1|1.00),(LOC109094368|LOC109094368|4|1.00),(LOC109100245|LOC109100245|9|0.89),(LOC122146593|LOC122146593|1|1.00),(LOC109095415|LOC109095415|1|1.00),(LOC122138786|LOC122138786|1|1.00),(LOC109052115|LOC109052115|2|1.00),(LOC109079158|LOC109079158|8|1.00),(LOC109061870|LOC109061870|1|1.00),(LOC122138707|LOC122138707|1|1.00),(LOC109090362|LOC109090362|1|1.00),(LOC109064567|LOC109064567|1|1.00),(LOC109077144|LOC109077144|1|1.00),(LOC109093640|LOC109093640|2|1.00),(LOC109057260|LOC109057260|1|1.00),(LOC109113423|LOC109113423|4|1.00),(LOC109086893|LOC109086893|2|1.00),(LOC109060930|LOC109060930|5|1.00),(LOC109094284|LOC109094284|1|1.00),(LOC109095492|LOC109095492|4|1.00),(LOC122134481|LOC122134481|6|1.00),(LOC109070447|LOC109070447|1|1.00),(LOC109088128|LOC109088128|1|1.00),(si:dkey-26i13.8|si:dkey-26i13.8|1|1.00),(LOC109091060|LOC109091060|5|1.00),(xpa|xpa|1|1.00),(LOC122145905|LOC122145905|1|1.00),(LOC109051882|LOC109051882|1|1.00),(LOC109060387|LOC109060387|3|1.00),(LOC109070584|LOC109070584|1|1.00),(LOC109074885|LOC109074885|2|1.00),(grk4|grk4|4|1.00),(LOC109078237|LOC109078237|1|1.00),(LOC109052585|LOC109052585|2|0.50),(LOC109064239|LOC109064239|2|1.00),(LOC109095406|LOC109095406|2|1.00),(LOC109095481|LOC109095481|1|1.00),(LOC109077732|LOC109077732|1|1.00),(LOC122146383|LOC122146383|1|1.00),(LOC109074794|LOC109074794|1|1.00),(LOC109071105|LOC109071105|1|1.00),(LOC109068179|LOC109068179|2|0.50),(LOC122138813|LOC122138813|1|1.00),(LOC109060541|LOC109060541|2|1.00),(LOC109100636|LOC109100636|2|1.00),(LOC109052588|LOC109052588|4|1.00),(LOC109108968|LOC109108968|1|1.00),(LOC109098990|LOC109098990|4|1.00),(LOC109066378|LOC109066378|1|1.00),(LOC109064236|LOC109064236|1|1.00),(ndfip2|ndfip2|1|1.00),(tspan5b|tspan5b|1|1.00),(LOC109094653|LOC109094653|3|1.00),(LOC109066109|LOC109066109|2|1.00),(LOC109081865|LOC109081865|2|1.00),(LOC109051716|LOC109051716|3|1.00),(LOC109054415|LOC109054415|1|1.00),(LOC109066750|LOC109066750|1|1.00),(LOC109051902|LOC109051902|2|1.00),(LOC109063029|LOC109063029|3|1.00),(LOC109062975|LOC109062975|1|1.00),(LOC109108976|LOC109108976|1|1.00),(LOC109068183|LOC109068183|1|1.00),(LOC109077898|LOC109077898|2|1.00),(LOC109099299|LOC109099299|2|1.00),(LOC109084847|LOC109084847|1|1.00),(LOC109051881|LOC109051881|1|1.00),(LOC109064193|LOC109064193|4|1.00),(LOC109064231|LOC109064231|1|1.00),(LOC109072040|LOC109072040|6|1.00),(slc24a2|slc24a2|2|1.00),(LOC122139615|LOC122139615|1|1.00),(LOC109063886|LOC109063886|1|1.00),(LOC109061405|LOC109061405|1|1.00),(LOC109081871|LOC109081871|4|1.00),(LOC109108981|LOC109108981|2|1.00),(LOC109085056|LOC109085056|2|1.00),(LOC109049467|LOC109049467|3|1.00),(LOC109082538|LOC109082538|2|1.00),(LOC109068182|LOC109068182|2|1.00),(LOC109086150|LOC109086150|1|1.00),(LOC109071097|LOC109071097|1|1.00),(LOC109098823|LOC109098823|1|1.00),(LOC109108964|LOC109108964|1|1.00),(LOC109068012|LOC109068012|1|1.00),(LOC109064184|LOC109064184|1|1.00),(LOC109068184|LOC109068184|1|1.00),(LOC122146602|LOC122146602|1|1.00),(LOC109081867|LOC109081867|4|1.00),(LOC109068102|LOC109068102|3|1.00),(LOC109108622|LOC109108622|1|1.00),(LOC109088130|LOC109088130|1|1.00),(LOC109069351|LOC109069351|1|1.00),(LOC109078080|LOC109078080|1|1.00),(LOC109074882|LOC109074882|1|1.00),(LOC109052551|LOC109052551|1|1.00),(LOC109108963|LOC109108963|1|1.00),(LOC109095694|LOC109095694|1|1.00),(LOC109088132|LOC109088132|1|1.00),(LOC109063976|LOC109063976|1|1.00),(LOC122138844|LOC122138844|1|1.00),(LOC109108971|LOC109108971|1|1.00),(LOC109074881|LOC109074881|1|1.00),(LOC109086113|LOC109086113|2|1.00),(LOC109090659|LOC109090659|1|1.00),(LOC109069582|LOC109069582|2|1.00),(LOC109068180|LOC109068180|1|1.00),(LOC109044867|LOC109044867|2|1.00),(LOC109111928|LOC109111928|2|1.00),(LOC109062389|LOC109062389|1|1.00),(LOC109074737|LOC109074737|2|1.00),(LOC109085054|LOC109085054|1|1.00),(LOC109100816|LOC109100816|7|1.00),(LOC109079183|LOC109079183|1|1.00),(LOC109083127|LOC109083127|2|1.00),(LOC109072834|LOC109072834|3|1.00),(LOC122140343|LOC122140343|1|1.00),(LOC109096812|LOC109096812|4|1.00),(LOC109057259|LOC109057259|1|1.00) | GT:GL:GQ:FT:RCL:RC:RCR:RDCN:DR:DV:RR:RV | 1/1:-17.8994,-1.50456,0:15:PASS:25777:47572:22800:2:0:3:0:5 |
| NC_056572.1 | 6526919 | DEL00000158 | A | <DEL> | 456.0 | PASS | PRECISE;SVTYPE=DEL;SVMETHOD=EMBL.DELLYv1.1.6;END=6528440;PE=4;MAPQ=60;CT=3to5;CIPOS=-4,4;CIEND=-4,4;SRMAPQ=60;INSLEN=0;HOMLEN=4;SR=4;SRQ=0.987013;CONSENSUS=ATTTAGCTTTGATTTGAAAGAAAACAGCGAATCCTTGTGGCGCGGATGATTCAGAAGAGCATATGCAGCACACAGCATGTTATTAGCTTGTATATATATATATAATTGTTGTTCAGTTAGGGCCACTGTAAGAAAAAATATGTGGAAAAACTGA;CE=1.91445;AC=2;AN=2;ANN=<DEL>|upstream_gene_variant|MODIFIER|LOC109108964|LOC109108964|transcript|XM_042726944.1|protein_coding||c.-2710_-1190del|||||983|,<DEL>|upstream_gene_variant|MODIFIER|LOC109108968|LOC109108968|transcript|XM_019122085.2|protein_coding||c.-2694_-1174del|||||2468|,<DEL>|intergenic_region|MODIFIER|LOC109108964-LOC109108968|LOC109108964-LOC109108968|intergenic_region|LOC109108964-LOC109108968|||n.6526920_6528440del|||||| | GT:GL:GQ:FT:RCL:RC:RCR:RDCN:DR:DV:RR:RV | 1/1:-17.6994,-1.50451,0:15:PASS:2:1:9:0:0:4:0:5 |
| NC_056572.1 | 6719676 | DEL00000168 | A | <DEL> | 600.0 | PASS | PRECISE;SVTYPE=DEL;SVMETHOD=EMBL.DELLYv1.1.6;END=6721197;PE=4;MAPQ=60;CT=3to5;CIPOS=-6,6;CIEND=-6,6;SRMAPQ=60;INSLEN=0;HOMLEN=6;SR=6;SRQ=0.994845;CONSENSUS=ATTTAATGGATCCTTGCTGAATGAAAGTATTAATTTCTTTAAAAAACCCAATACCAAATTTTGAACGGTAGTGTAAATATATAGCATATCCAAAGCTCACTGAAACTACAGAGATGGAGGTACACAGATACACTCATACCTTTTTGCTTGCTTCAAAAGATCTGGACCGTTTCTTGTGCTGGTGTCTCTCAGGC;CE=1.94703;AC=2;AN=2;ANN=<DEL>|intron_variant|MODIFIER|si:dkey-26i13.8|si:dkey-26i13.8|transcript|XM_042728781.1|protein_coding|18/20|c.2550+60_2550+1580del|||||| | GT:GL:GQ:FT:RCL:RC:RCR:RDCN:DR:DV:RR:RV | 1/1:-17.7994,-1.50454,0:15:PASS:5:0:12:0:0:4:0:5 |
表3.5.1 SV注释后变异信息记录文件示例
- #CHROM: 变异所在的染色体或者参考序列的名称。
- POS: 变异在染色体上的碱基位置。
- ID: 变异ID。通常为dbSNP数据库中的rs编号,如果没有则为“.”。
- REF: 参考基因组序列。该位点在参考基因组上的碱基序列。
- ALT: 变异序列。样本中的碱基序列,如与参考基因组一致则显示为“.”。
- QUAL: 质量得分。变异检测的质量分数。
- FILTER: 过滤状态。表示变异是否通过了质控。
- INFO: 变异额外信息。其中ANN存储了注释信息
- FORMAT: 变异存储格式。定义样本列中数据的顺序。
- Sample: 样本名。
以下为注释信息统计。表格右上角的检索框可用于筛选包含检索内容的条目,即仅显示包含输入关键词的行
| Type | Sample1 | Sample2 | Sample3 | Sample4 | Sample5 |
|---|---|---|---|---|---|
| 3 Prime UTR Variant | 5 | 14 | 9 | 17 | 15 |
| 5 Prime UTR Truncation | 3 | 9 | 8 | 5 | 2 |
| 5 Prime UTR Variant | 23 | 14 | 18 | 19 | 5 |
| Bidirectional Gene Fusion | 137 | 126 | 78 | 98 | 45 |
| Chromosome Number Variation | 10 | 14 | 9 | 5 | 10 |
| Conservative Inframe Deletion | 3 | 5 | 2 | 3 | 6 |
| Disruptive Inframe Deletion | 1 | 0 | 2 | 2 | 1 |
| Downstream Gene Variant | 367 | 386 | 441 | 385 | 375 |
| Duplication | 17219 | 8170 | 12580 | 8115 | 803 |
| Exon Loss Variant | 428 | 449 | 289 | 111 | 337 |
| Feature Ablation | 1053 | 728 | 1117 | 845 | 1062 |
| Feature Fusion | 184 | 176 | 213 | 338 | 193 |
| Frameshift Variant | 332 | 111 | 151 | 82 | 96 |
| Gene Fusion | 532 | 241 | 476 | 191 | 624 |
| Intergenic Region | 423 | 425 | 502 | 417 | 418 |
| Intragenic Variant | 546 | 353 | 403 | 348 | 166 |
| Intron Variant | 1123 | 1091 | 1115 | 1147 | 1038 |
| Inversion | 6680 | 7100 | 8859 | 5660 | 9973 |
| Non Coding Transcript Exon Variant | 11 | 14 | 12 | 2 | 10 |
| Non Coding Transcript Variant | 9 | 19 | 28 | 27 | 17 |
| Splice Acceptor Variant | 21 | 20 | 26 | 5 | 10 |
| Splice Donor Variant | 32 | 26 | 28 | 22 | 14 |
| Splice Region Variant | 70 | 76 | 64 | 55 | 41 |
| Stop Gained | 11 | 15 | 9 | 4 | 4 |
| Stop Lost | 6 | 2 | 2 | 1 | 1 |
| Transcript Ablation | 4159 | 3467 | 5257 | 3634 | 4895 |
| Upstream Gene Variant | 452 | 398 | 533 | 427 | 375 |
| 3 Prime UTR Truncation | 0 | 1 | 2 | 0 | 1 |
| Exon Region | 0 | 1 | 8 | 4 | 1 |
| Start Lost | 0 | 4 | 3 | 4 | 0 |
表3.5.2 SV变异信息统计(按对基因的影响预测统计)
- Type: 变异产生了哪些影响,具体释义请参照如下页面的functional-class一节。https://pcingola.github.io/SnpEff/snpeff/inputoutput
| Type | Sample1 | Sample2 | Sample3 | Sample4 | Sample5 |
|---|---|---|---|---|---|
| Chromosome | 49 | 43 | 41 | 31 | 37 |
| Downstream | 367 | 386 | 440 | 384 | 375 |
| Exon | 834 | 692 | 839 | 632 | 459 |
| Gene | 7721 | 5574 | 7442 | 4869 | 5996 |
| Intergenic | 607 | 601 | 715 | 755 | 611 |
| Intron | 1076 | 1053 | 1072 | 1121 | 1017 |
| Splice Site Acceptor | 8 | 8 | 13 | 4 | 6 |
| Splice Site Donor | 22 | 18 | 13 | 17 | 10 |
| Splice Site Region | 8 | 25 | 15 | 19 | 18 |
| Transcript | 22191 | 14374 | 20791 | 13513 | 11459 |
| Upstream | 452 | 398 | 533 | 427 | 375 |
| UTR 3 Prime | 5 | 9 | 9 | 13 | 14 |
| UTR 5 Prime | 7 | 7 | 14 | 11 | 3 |
表3.5.3 SV变异信息统计(按区域统计)
- Type: 变异发生在哪些区域,具体释义请参照如下页面的Variant annotaiton details一节。https://pcingola.github.io/SnpEff/snpeff/inputoutput
| Sample ID | BND | INV | DEL | DUP |
|---|---|---|---|---|
| Sample1 | 166 | 55 | 771 | 52 |
| Sample2 | 139 | 43 | 749 | 38 |
| Sample3 | 141 | 42 | 778 | 54 |
| Sample4 | 142 | 34 | 763 | 43 |
| Sample5 | 154 | 50 | 764 | 36 |
表3.5.4 SV变异类型统计
- Sample ID: 样本ID
- BND: 跨染色体移位变异
- DEL: 大片段缺失变异
- DUP: 复制变异
- INV: 倒位变异
- 图3.5.1与3.5.2 展示了每个样本中SV变异的预测饼图, 其中3.5.1是按照变异对基因的影响进行统计,3.5.2是按照变异位点所在区域进行统计。 由于参考基因组注释文件中对单个基因会标记多个transcript(转录本), 所以预测的影响会比变异记录文件具有更多条目数。
- 图3.5.3 展示了每个样本中SV变异的类型统计, 其中INS代表大片段插入变异,DEL代表大片段缺失变异,DUP代表复制变异,INV代表倒位变异, BND代表跨染色体移位变异。 其中,X轴表示每个样本,Y轴表示碱基变异情况在样本中出现的次数。 拖拽滑条可以自行选择展示范围。
- 图3.5.4 展示了每个样本中不同类型的SV变异的长度分布, 其中,X轴表示每个样本,Y轴表示碱基该类型变异的长度分布。 拖拽滑条可以自行选择展示范围。
3.6 CNV检测与注释
流程结果
├── 06.CNV (进入文件目录)
│ ├── Sample1
│ │ ├── 表3.6.4_CNV长度统计_Sample1.csv (因数据过多,未列入报告)
│ │ ├── 图3.6.2_样本CNV效应类型占比_Sample1.png
│ │ └── 图3.6.3_样本CNV发生位点占比_Sample1.png
│ ├── ...
│ ├── 表3.6.2_CNV变异信息统计(按对基因的影响预测统计).csv
│ ├── 表3.6.3_CNV变异信息统计(按区域统计).csv
│ └── 图3.6.4_样本CNV长度分布箱线图.png
└── ...
CNV(Copy Number Variants)指的是DNA片段在基因组中的拷贝数上发生的变异。这个片段的长度不一,可以覆盖一个基因乃至上兆的碱基。 虽然检测结构性变异的工具通常也能用来检测拷贝数变异,但由于拷贝数变异的特殊性质(如成片段加倍),使得它们在检测CNV上的精确性不如专门被设计于CNV检测的工具。 为了更准确地分析拷贝数变异,本次分析采用了CNVnator[6](参数:窗口大小 10000bp)对所有样本的比对数据进行变异检测。 得到的CNV结果经过后处理和过滤,以确保结果的准确性和可靠性。
CNVnator的流程介绍如下:
- CNVnator是基于测序深度(Read Depth)检测拷贝数变异(CNVs)的工具
- 该算法的计算步骤如下:
- 基因组分窗:参考基因组将首先按固定大小分割为非重叠的窗口(Bin)。窗口的大小被称作分辨率,影响检测拷贝数变异的精度。
- 读取深度数据:将比对数据(BAM文件)排列至每个窗口上,进而计算每个窗口的测序深度。此处假设测序深度与该窗口的拷贝数成正比。
- 深度数据标准化:对每个窗口的测序深度进行标准化,以减少GC含量与平台误差等因素对测序获得的深度的影响。
- 识别测序深度的变化信号:计算全基因组或特定染色体上所有窗口的平均测序深度及其方差,并评估每个窗口的测序深度是否显著偏离均值。若是,则表明该窗口可能存在拷贝数变异。
- 分段合并:获得全局异常测序深度的信号后,相邻且测序深度信号相似的窗口将被合并为单一的拷贝数变异区域,并依据这一信号判断变异类型。
以下为结果文件示例
| #CHROM | POS | ID | REF | ALT | QUAL | FILTER | INFO | FORMAT | Sample1 |
|---|---|---|---|---|---|---|---|---|---|
| NC_056572.1 | 3230001 | Sample1_CNVnator_DEL_1 | N | <DEL> | . | PASS | END=3540000;SVTYPE=DEL;SVLEN=-310000;IMPRECISE;natorRD=0.24974;natorP1=5.14105e-13;natorP2=2.30189e-29;natorP3=5.14105e-13;natorP4=2.30189e-29;natorQ0=0.086087;AC=2;AN=2;ANN=<DEL>|feature_ablation|HIGH|LOC109071461&LOC109080069&LOC122146585&LOC122146715&LOC122146719|LOC109071461&LOC109080069&LOC122146585&LOC122146715&LOC122146719|gene_variant|LOC109080069|||n.3540000_3230002del||||||,<DEL>|transcript_ablation|HIGH|LOC122146719|LOC122146719|transcript|XR_006161239.1|pseudogene|2/4|n.-266693_*14837del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122146715|LOC122146715|transcript|XR_006161234.1|pseudogene|2/3|n.-223965_*80986del|p.0?|||||,<DEL>|transcript_ablation|HIGH|LOC122146715|LOC122146715|transcript|XR_006161236.1|pseudogene|2/3|n.-223965_*83099del|p.0?|||||,<DEL>|exon_loss_variant|HIGH|LOC109080069|LOC109080069|transcript|XM_042766177.1|protein_coding|12/12|c.430+238_*305939del||||||,<DEL>|exon_loss_variant|HIGH|LOC109080069|LOC109080069|transcript|XM_042766177.1|protein_coding|11/12|c.430+238_*305939del||||||,<DEL>|exon_loss_variant|HIGH|LOC109080069|LOC109080069|transcript|XM_042766177.1|protein_coding|10/12|c.430+238_*305939del||||||,<DEL>|exon_loss_variant|HIGH|LOC109080069|LOC109080069|transcript|XM_042766177.1|protein_coding|9/12|c.430+238_*305939del||||||,<DEL>|exon_loss_variant|HIGH|LOC109080069|LOC109080069|transcript|XM_042766177.1|protein_coding|8/12|c.430+238_*305939del||||||,<DEL>|exon_loss_variant|HIGH|LOC109080069|LOC109080069|transcript|XM_042766177.1|protein_coding|7/12|c.430+238_*305939del||||||,<DEL>|exon_loss_variant|HIGH|LOC109080069|LOC109080069|transcript|XM_042766177.1|protein_coding|6/12|c.430+238_*305939del||||||,<DEL>|exon_loss_variant|HIGH|LOC109080069|LOC109080069|transcript|XM_042766177.1|protein_coding|5/12|c.430+238_*305939del||||||,<DEL>|exon_loss_variant|HIGH|LOC109080069|LOC109080069|transcript|XM_042766177.1|protein_coding|4/12|c.430+238_*305939del||||||,<DEL>|exon_loss_variant|HIGH|LOC109080069|LOC109080069|transcript|XM_042766177.1|protein_coding|3/12|c.430+238_*305939del||||||,<DEL>|upstream_gene_variant|MODIFIER|LOC122146719|LOC122146719|transcript|XR_006161239.1|pseudogene|2/4|n.-266693_*14837del|||||0|,<DEL>|upstream_gene_variant|MODIFIER|LOC122146715|LOC122146715|transcript|XR_006161234.1|pseudogene|2/3|n.-223965_*80986del|||||0|,<DEL>|upstream_gene_variant|MODIFIER|LOC122146715|LOC122146715|transcript|XR_006161236.1|pseudogene|2/3|n.-223965_*83099del|||||0|,<DEL>|downstream_gene_variant|MODIFIER|LOC122146719|LOC122146719|transcript|XR_006161239.1|pseudogene|2/4|n.-266693_*14837del|||||14837|,<DEL>|downstream_gene_variant|MODIFIER|LOC122146715|LOC122146715|transcript|XR_006161234.1|pseudogene|2/3|n.-223965_*80986del|||||80986|,<DEL>|downstream_gene_variant|MODIFIER|LOC109080069|LOC109080069|transcript|XM_042766177.1|protein_coding|3/12|c.430+238_*305939del|||||304223|,<DEL>|downstream_gene_variant|MODIFIER|LOC122146715|LOC122146715|transcript|XR_006161236.1|pseudogene|2/3|n.-223965_*83099del|||||83099|,<DEL>|intragenic_variant|MODIFIER|LOC109071461|LOC109071461|gene_variant|LOC109071461|||n.3230002_3540000del||||||,<DEL>|intragenic_variant|MODIFIER|LOC122146585|LOC122146585|gene_variant|LOC122146585|||n.3230002_3540000del||||||,<DEL>|intergenic_region|MODIFIER|LOC109091551-LOC122146719|LOC109091551-LOC122146719|intergenic_region|LOC109091551-LOC122146719|||n.3230002_3540000del||||||,<DEL>|intergenic_region|MODIFIER|LOC122146719-LOC122146715|LOC122146719-LOC122146715|intergenic_region|LOC122146719-LOC122146715|||n.3230002_3540000del||||||,<DEL>|intergenic_region|MODIFIER|LOC122146715-LOC109071461|LOC122146715-LOC109071461|intergenic_region|LOC122146715-LOC109071461|||n.3230002_3540000del||||||,<DEL>|intergenic_region|MODIFIER|LOC109071461-LOC122146585|LOC109071461-LOC122146585|intergenic_region|LOC109071461-LOC122146585|||n.3230002_3540000del||||||,<DEL>|intergenic_region|MODIFIER|LOC122146585-LOC109080069|LOC122146585-LOC109080069|intergenic_region|LOC122146585-LOC109080069|||n.3230002_3540000del||||||;LOF=(LOC109080069|LOC109080069|1|1.00) | GT:CN | 1/1:0 |
| NC_056572.1 | 5930001 | Sample1_CNVnator_DEL_2 | N | <DEL> | . | PASS | END=6010000;SVTYPE=DEL;SVLEN=-80000;IMPRECISE;natorRD=0.373443;natorP1=1.08736;natorP2=5.20863e-09;natorP3=1.08736;natorP4=5.20863e-09;natorQ0=0.0642361;AC=1;AN=2;ANN=<DEL>|intergenic_region|MODIFIER|LOC109113423-LOC122146595|LOC109113423-LOC122146595|intergenic_region|LOC109113423-LOC122146595|||n.5930002_6010000del|||||| | GT:CN | 0/1:1 |
| NC_056572.1 | 6020001 | Sample1_CNVnator_DEL_3 | N | <DEL> | . | PASS | END=6080000;SVTYPE=DEL;SVLEN=-60000;IMPRECISE;natorRD=0.0814445;natorP1=0.0247397;natorP2=4.28664e-25;natorP3=0.0247397;natorP4=4.28664e-25;natorQ0=0.721053;AC=2;AN=2;ANN=<DEL>|intragenic_variant|MODIFIER|LOC122146595|LOC122146595|gene_variant|LOC122146595|||n.6020002_6080000del||||||,<DEL>|intergenic_region|MODIFIER|LOC109113423-LOC122146595|LOC109113423-LOC122146595|intergenic_region|LOC109113423-LOC122146595|||n.6020002_6080000del||||||,<DEL>|intergenic_region|MODIFIER|LOC122146595-LOC109083632|LOC122146595-LOC109083632|intergenic_region|LOC122146595-LOC109083632|||n.6020002_6080000del|||||| | GT:CN | 1/1:0 |
| NC_056572.1 | 10530001 | Sample1_CNVnator_DEL_4 | N | <DEL> | . | PASS | END=10630000;SVTYPE=DEL;SVLEN=-100000;IMPRECISE;natorRD=0.403113;natorP1=0.257973;natorP2=0.00425243;natorP3=0.257973;natorP4=0.00425243;natorQ0=0.199688;AC=1;AN=2;ANN=<DEL>|intergenic_region|MODIFIER|LOC109074737-LOC122140423|LOC109074737-LOC122140423|intergenic_region|LOC109074737-LOC122140423|||n.10530002_10630000del|||||| | GT:CN | 0/1:1 |
| NC_056572.1 | 12050001 | Sample1_CNVnator_DEL_5 | N | <DEL> | . | PASS | END=12440000;SVTYPE=DEL;SVLEN=-390000;IMPRECISE;natorRD=0.230934;natorP1=4.08647e-13;natorP2=2.6737e-15;natorP3=4.08647e-13;natorP4=2.6737e-15;natorQ0=0.104312;AC=2;AN=2;ANN=<DEL>|downstream_gene_variant|MODIFIER|LOC109062975|LOC109062975|transcript|XM_019080033.2|protein_coding||c.*3836_*393834del|||||3831|,<DEL>|intergenic_region|MODIFIER|LOC109062975-LOC109112091|LOC109062975-LOC109112091|intergenic_region|LOC109062975-LOC109112091|||n.12050002_12440000del|||||| | GT:CN | 1/1:0 |
表3.6.1 CNV注释后变异信息记录文件示例
- #CHROM: 变异所在的染色体或者参考序列的名称。
- POS: 变异在染色体上的碱基位置。
- ID: 变异ID。通常为dbSNP数据库中的rs编号,如果没有则为“.”。
- REF: 参考基因组序列。该位点在参考基因组上的碱基序列。
- ALT: 变异序列。样本中的碱基序列,如与参考基因组一致则显示为“.”。
- QUAL: 质量得分。变异检测的质量分数。
- FILTER: 过滤状态。表示变异是否通过了质控。
- INFO: 变异额外信息。其中ANN存储了注释信息
- FORMAT: 变异存储格式。定义样本列中数据的顺序。
- Sample: 样本名。
| Type | Sample1 | Sample2 | Sample3 | Sample4 | Sample5 |
|---|---|---|---|---|---|
| 3 Prime UTR Truncation | 88 | 80 | 94 | 97 | 101 |
| 3 Prime UTR Variant | 34 | 36 | 21 | 46 | 38 |
| 5 Prime UTR Truncation | 219 | 229 | 210 | 224 | 220 |
| 5 Prime UTR Variant | 24 | 24 | 24 | 14 | 17 |
| Bidirectional Gene Fusion | 232 | 213 | 198 | 184 | 252 |
| Chromosome Number Variation | 238 | 256 | 245 | 247 | 251 |
| Conservative Inframe Deletion | 52 | 76 | 63 | 52 | 75 |
| Disruptive Inframe Deletion | 3 | 2 | 2 | 0 | 4 |
| Downstream Gene Variant | 4097 | 4095 | 4101 | 3916 | 3947 |
| Duplication | 4587 | 4750 | 6351 | 4395 | 5832 |
| Exon Loss Variant | 6401 | 6338 | 5810 | 5193 | 6437 |
| Feature Ablation | 2953 | 3050 | 2911 | 2974 | 2970 |
| Frameshift Variant | 128 | 123 | 117 | 114 | 137 |
| Gene Fusion | 343 | 458 | 831 | 324 | 929 |
| Intergenic Region | 5655 | 5386 | 5518 | 5457 | 5507 |
| Intragenic Variant | 2884 | 2848 | 2843 | 2809 | 2846 |
| Intron Variant | 652 | 669 | 612 | 711 | 641 |
| Non Coding Transcript Exon Variant | 56 | 65 | 57 | 56 | 54 |
| Non Coding Transcript Variant | 147 | 156 | 142 | 180 | 156 |
| Splice Acceptor Variant | 133 | 150 | 143 | 160 | 142 |
| Splice Donor Variant | 286 | 309 | 276 | 301 | 292 |
| Splice Region Variant | 677 | 718 | 655 | 724 | 700 |
| Start Lost | 67 | 76 | 63 | 64 | 74 |
| Stop Gained | 46 | 37 | 138 | 38 | 128 |
| Stop Lost | 77 | 87 | 67 | 61 | 104 |
| Transcript Ablation | 6860 | 7007 | 7006 | 7279 | 7000 |
| Upstream Gene Variant | 4239 | 4152 | 4224 | 4014 | 4102 |
表3.6.2 CNV变异信息统计(按对基因的影响预测统计)
- Type: 变异产生了哪些影响,具体释义请参照如下页面的functional-class一节。https://pcingola.github.io/SnpEff/snpeff/inputoutput
| Type | Sample1 | Sample2 | Sample3 | Sample4 | Sample5 |
|---|---|---|---|---|---|
| Chromosome | 270 | 285 | 278 | 279 | 280 |
| Downstream | 4097 | 4093 | 4100 | 3915 | 3947 |
| Exon | 6904 | 7236 | 7425 | 5532 | 8350 |
| Gene | 4304 | 4497 | 4858 | 4277 | 4887 |
| Intergenic | 5655 | 5386 | 5518 | 5457 | 5507 |
| Intron | 259 | 243 | 218 | 280 | 233 |
| Splice Site Acceptor | 34 | 52 | 39 | 68 | 36 |
| Splice Site Donor | 245 | 255 | 239 | 249 | 245 |
| Splice Site Region | 135 | 147 | 129 | 151 | 147 |
| Transcript | 13041 | 12946 | 13649 | 13337 | 13045 |
| Upstream | 4239 | 4152 | 4224 | 4014 | 4102 |
| UTR 3 Prime | 29 | 25 | 19 | 35 | 36 |
| UTR 5 Prime | 19 | 17 | 13 | 9 | 10 |
表3.6.3 CNV变异信息统计(按区域统计)
- Type: 变异发生在哪些区域,具体释义请参照如下页面的Variant annotaiton details一节。https://pcingola.github.io/SnpEff/snpeff/inputoutput
| Sample ID | DEL | DUP |
|---|---|---|
| Sample1 | 2204 | 60 |
| Sample2 | 2146 | 63 |
| Sample3 | 2060 | 64 |
| Sample4 | 2139 | 64 |
| Sample5 | 2161 | 54 |
表3.6.4 CNV变异类型统计
- Sample ID: 样本ID
- DUP: 拷贝数增加变异
- DEL: 拷贝数减少变异
- 图3.6.1与3.5.2 展示了每个样本中CNV变异的预测饼图, 其中3.6.1是按照变异对基因的影响进行统计,3.6.2是按照变异位点所在区域进行统计。 由于参考基因组注释文件中对单个基因会标记多个transcript(转录本), 所以预测的影响会比变异记录文件具有更多条目数。
- 图3.6.3 展示了每个样本中CNV变异的长度分布箱线图。 其中,X轴表示每个样本,Y轴表示变异长度。 拖拽滑条可以自行选择展示范围。
3.7 CNV与SV的Circos图
流程结果
└── 07.Circos (进入文件目录)
├── 图3.7.1_CNV_SV_circos_Sample1.png
└── ...
图3.7.1 SV_CNV Circos图
- 图3.7.1 展示了每个样本中SV变异与CNV变异的Circos图, 图层从内向外依次为染色体长度刻度(单位为10Mb),拷贝数变异热度指示,染色体倒位(Inversion)位置指示 与染色体易位(Translocation)位置指示。
- 拷贝数变异热度指示中,数据点表示CNV变异发生位置,点所处纵坐标表示此位置拷贝数量。 由于通常情况下CNV检测的结果跨度区间较小,因此若非变异特别密集的区域,在图片上不会有非常清晰的展示
- 染色体倒位(Inversion)位置指示中,线条表示倒位发生的染色体区域, 鼠标指针移至对应线条上后可以显示倒位区域的具体信息。
- 染色体易位(Translocation)位置指示中,线条表示易位发生的染色体区域, 鼠标指针移至对应线条上后可以显示易位区域的具体信息。
四 分析所用软件的版本
| 软件 | 版本 |
|---|---|
| fastp | 0.23.4 |
| assembly-stat | 1.0.1 |
| Seqkit | 2.5.0 |
| BWA | 0.7.17 |
| sambamba | 1.0.0 |
| QualiMap | v.2.2.2a-1 |
| Freebayes | 1.3.6 |
| CNVnator | v0.4.1 |
| Delly | v1.1.6 |
| SNPeff | 5.1d |
| samtools | v1.7 |
| bcftools | v1.5 |
| vcftools | v0.1.16 |
五 参考文献
- [1] Chen, S., Zhou, Y., Chen, Y., & Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34(17), i884-i890.
- [2] Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997.
- [3] Garrison, E., & Marth, G. (2012). Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907.
- [4] Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., ... & Ruden, D. M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. fly, 6(2), 80-92.
- [5] Rausch, T., Zichner, T., Schlattl, A., Stütz, A. M., Benes, V., & Korbel, J. O. (2012). DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics, 28(18), i333-i339.
- [6] Abyzov, A., Urban, A. E., Snyder, M., & Gerstein, M. (2011). CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome research, 21(6), 974-984.
- [7] Okonechnikov, K., Conesa, A., & García-Alcalde, F. (2016). Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics, 32(2), 292-294.