Input data and parameters
Input
| Analysis date: | Wed Apr 27 19:35:51 CST 2022 |
| BAM file: | /dev/stdin |
| Counting algorithm: | uniquely-mapped-reads |
| GTF file: | /home/bayegy/Databases/transcriptome/references/mmu/GCF_000001635.27_GRCm39_genomic.gtf |
| Number of bases for 5'-3' bias computation: | 100 |
| Number of transcripts for 5'-3' bias computation: | 1,000 |
| Paired-end sequencing: | yes |
| Protocol: | non-strand-specific |
| Sorting performed: | no |
Summary
Reads alignment
| Number of mapped reads (left/right): | 19,745,458 / 19,495,300 |
| Number of aligned pairs (without duplicates): | 17,454,061 |
| Total number of alignments: | 41,768,527 |
| Number of secondary alignments: | 2,527,769 |
| Number of non-unique alignments: | 3,996,225 |
| Aligned to genes: | 33,662,690 |
| Ambiguous alignments: | 321,407 |
| No feature assigned: | 3,787,980 |
| Missing chromosome in annotation: | 225 |
| Not aligned: | 1,565,642 |
| Strand specificity estimation (fwd/rev): | 0.5 / 0.5 |
Reads genomic origin
| Exonic: | 33,662,690 / 89.89% |
| Intronic: | 2,688,909 / 7.18% |
| Intergenic: | 1,099,071 / 2.93% |
| Intronic/intergenic overlapping exon: | 883,622 / 2.36% |
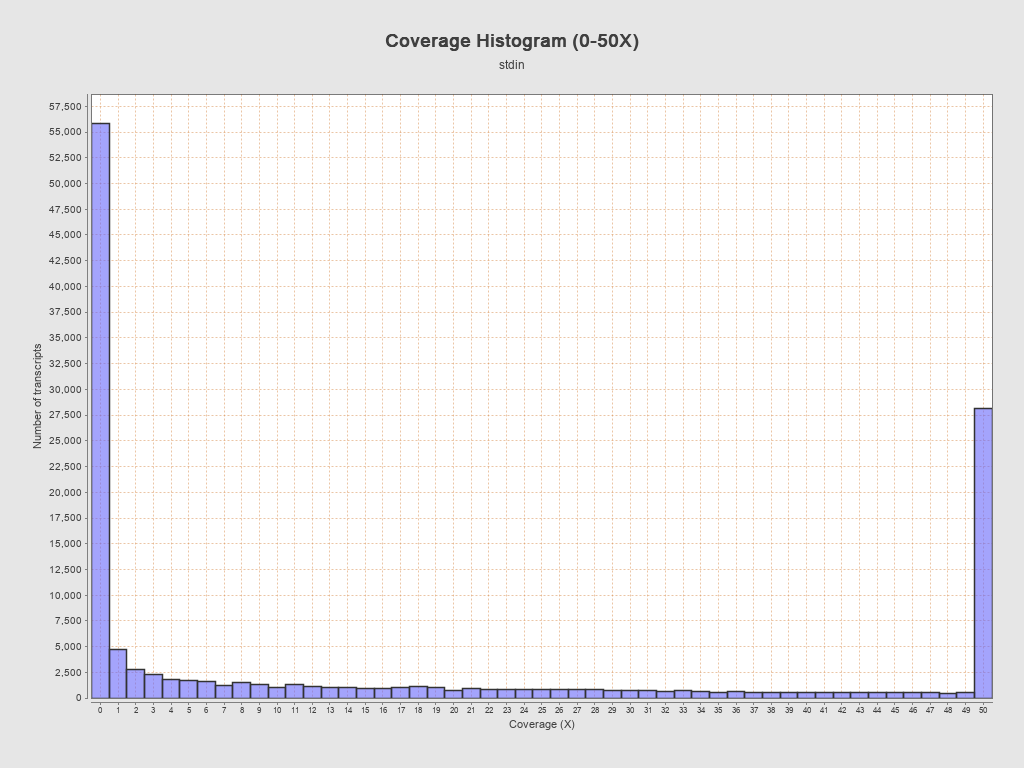
Transcript coverage profile
| 5' bias: | 0.68 |
| 3' bias: | 0.27 |
| 5'-3' bias: | 1.52 |
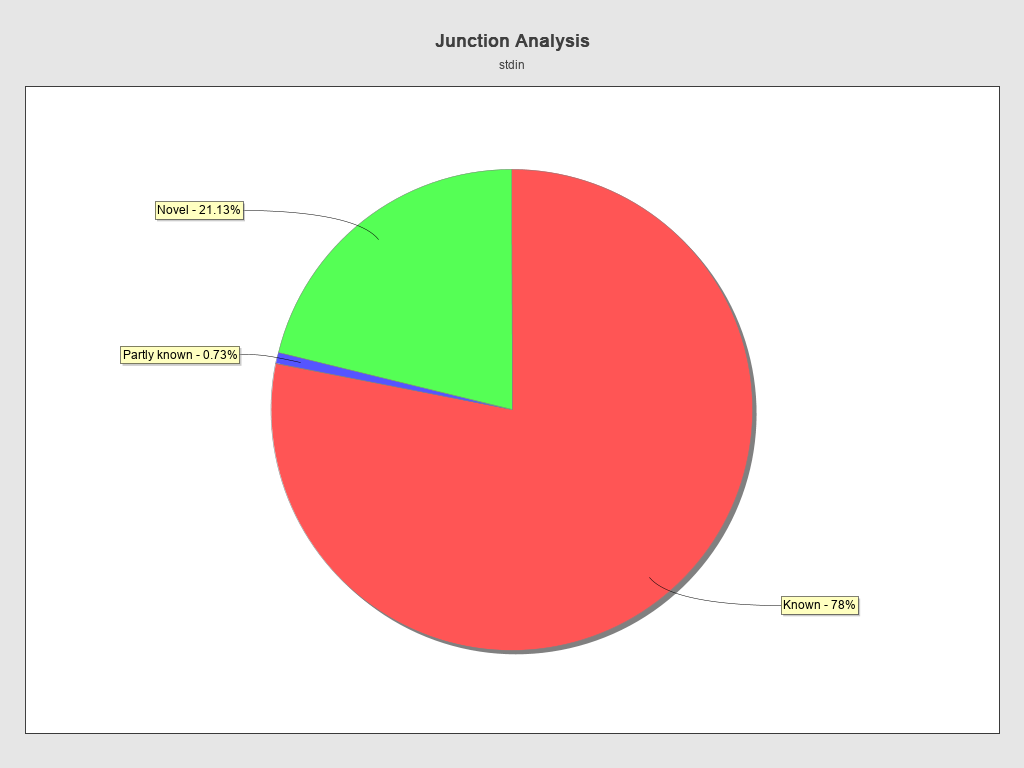
Junction analysis
| Reads at junctions: | 21,421,630 |
| AGGT | 4.43% |
| ACCT | 4.32% |
| AGGA | 3.51% |
| AGGG | 3.14% |
| CCCT | 2.93% |
| TCCT | 2.91% |
| AGCT | 2.82% |
| AGAT | 2.55% |
| AGAA | 2.47% |
| ATCT | 2.36% |
| AGGC | 2.35% |


.png)
.png)
.png)