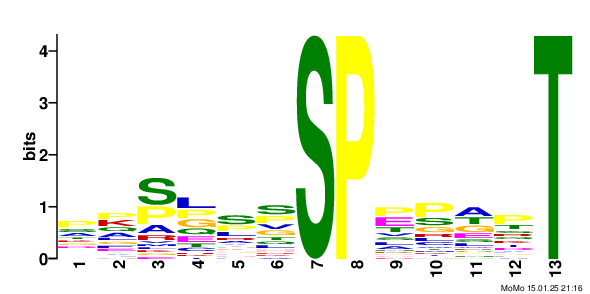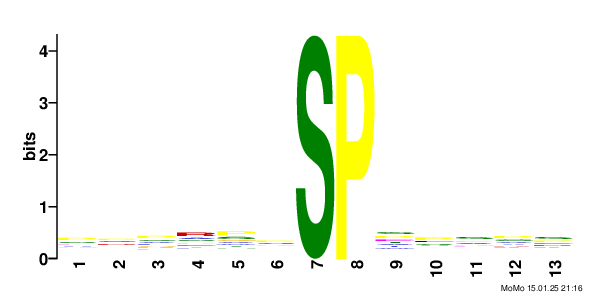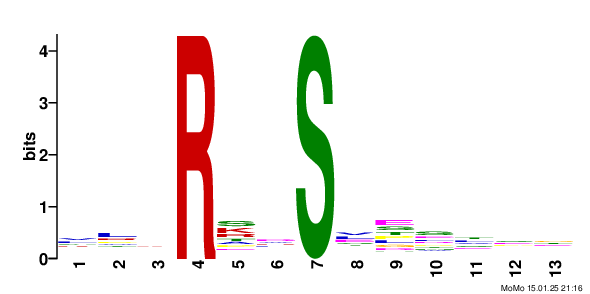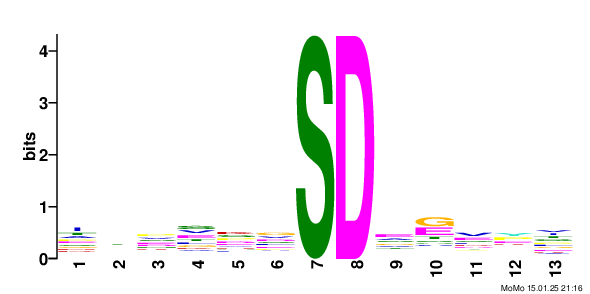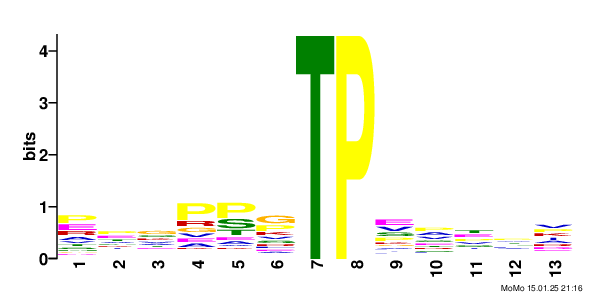MoMo Results
Modification Motifs
MoMo version 5.0.2 (Release date: Thu Aug 30 14:26:08 2018 -0700)
For further information on how to interpret these results please access http://meme-suite.org/doc/momo-output-format.html .http://meme-suite.org .
If you use MoMo in your research, please cite the following paper:[full text]
Peptide Preprocessing
Algorithm: motif-x
Number of Mods: 1465
Number of Mod Types: 3
Number of Mods Passing Filters: 1454
Number of Mod Types Passing Filters: 3
Results motif: xxxxxx_S79.9_PxxxxT score: 64.06 fg: 26/1062 bg: 2/1062 fg/bg: 13.0 unadjusted p -value: 1.3e-006 tests: 243 adjusted p -value: 3.2e-004 Show/Hide Motif Occurrences
MQSKPQSPVIQAT
GGSGRGSPSGGST
AEGIPPSPPGAGT
DKRLKLSPEWGPT
FLSEPSSPGRSKT
PASPAPSPVPSPT
NSSTQSSPEMPTT
EYPLSQSPNCGIT
DDAERRSPTPERT
GAALSPSPDCRVT
IFECLVSPSTAIT
PGKGVTSPLHTDT
TQPLSHSPKRTPT
KFPQGPSPEGLST
PAPQPHSPPSTKT
AKLPSSSPPLEPT
PKMPDKSPEPVDT
SPARSGSPAPETT
SPVQCVSPELALT
PPPPTASPHSNRT
TLSEELSPPSHQT
RPRGAGSPTPLPT
SESLLTSPPKACT
YVSCLSSPLSGPT
KKSTSSSPAPTQT
AQPGHVSPTPATT
motif: xxxxxx_S79.9_Pxxxxx score: 44.37 fg: 291/1062 bg: 110/1062 fg/bg: 2.6 unadjusted p -value: 2.1e-024 tests: 231 adjusted p -value: 4.8e-022 Show/Hide Motif Occurrences
LGGRPGSPRRTTP
QPRDSASPSTSSF
IQATAGSPKFSRL
SYGAVTSPTSTLE
KIYRSQSPHYFQS
YTAGSASPTPTFK
DENVLDSPKQRRS
PSPVTHSPLLHPR
AGGPGISPLASKH
RLFRTKSPAELSE
AKSLPPSPVTHSP
TKTPSASPRHGRS
LQGPPDSPSLGVA
PVTTGPSPEACKE
PFPTQVSPLLGSE
GVPLRTSPSLTPW
SAGPPASPGCVPR
KDDTPLSPSEKFK
DAAVPASPQSAGP
RCRVGGSPQPAVS
GELYMQSPALRAR
PKLGPSSPAHSGA
YTIRVSSPFGPQE
VQSMDVSPVSKEE
EESYPQSPRRGSE
HFVEPLSPTGVPG
PSTHPRSPSDPGG
PQMPRTSPQAPGL
ISHDRDSPPPPPP
VLADRESPEQPFM
SDLEDSSPRNSQE
RRSSQPSPTTVPA
EPAAPASPAPSPV
NPTGQISPQPAPP
KPGAGGSPALARR
LPPKPPSPRGTTA
APSPVPSPTPAQP
FRDRQRSPIALPV
PPPVAGSPRLPGG
VRDSCSSPPSLNY
VTKGGYSPQEGGD
NPRTVPSPPISPQ
HVTLPSSPRSNTP
GSTLRTSPADHGG
ELASPVSPELRQK
PPPLESSPRVKSP
DQKELASPVSPEL
PLTVPVSPKFSTR
SPPSPASPTSLNQ
PVSRSISPSPSAI
SKIPALSPSSGKS
LELSEDSPNSEQE
DKIGGKSPPPPPP
GDCKPTSPSLPAS
DRALQTSPQPVEG
SAPSATSPFYKAC
ATLQLGSPEASQS
ECKITGSPEIQVV
MKDRIVSPDLQLD
IPPKPESPPLEVP
QFIKKPSPVLVLR
ELGFSASPPSRSP
ASPPSRSPPRFEL
GELLRPSPTCEIK
GEPLTASPDCEII
LMLQSISPSDAGE
CKLTWFSPEDDGG
VAKKTPSPIEAER
RSPIRMSPAMSPA
RMSPAMSPARMSP
MSPARMSPARMSP
NAAGAISPPSEPS
PPARPPSPPKEDV
QLARQQSPSPIRH
ARQQSPSPIRHSP
RRRRSLSPTYIEL
ARRRTPSPDYDLY
DPVFLPSPPSKPK
RAEPTPSPQFPFA
DPFTTPSPPTSLE
VRAPTPSPVRSVS
SPVRSVSPAGRIS
PLEDGGSPIKAYV
EIPEPPSPEDLEI
SHPKAVSPTETKP
KDGAALSPSPDCR
GEEIVPSPKHSVK
PLKDGGSPILGYI
PDRDGGSPITGYL
PEKDGGSPITNYI
VATAQWSPLSTTS
LIIVPASPSDSGE
ITQREESPPPAVP
PLGPPTSPERLTY
ISEPLTSPKMLAK
TSIVKGSPPFTVS
PSPIRHSPSPVRH
PIRHSPSPVRHVR
PKIADASPDEGWK
PERIELSPSMEAP
ECKIGGSPEIKVL
CKVAGSSPISIAW
SLPSETSPLVRAE
SPKRVKSPEPVTS
PRSDGGSPIQGYI
PKSDGGSPITGYY
PRSLSRSPIRMSP
PEPRVKSPETVKS
SPETVKSPKRVKS
ERTRPRSPSPVSS
TRPRSPSPVSSER
PKPKSRSPTPPSI
EVSGEPSPEIEWF
SVDLEWSPPLKDG
PIRSVKSPLLIRK
VQKTAESPEAKKQ
KRPRTASPHFTVS
NVRVLDSPSAPVN
WQRVNTSPISGRE
GNEIISSPKCQSS
NSAGLSSPSDPSK
AGLSNPSPSSDPI
EPVETDSPVEARS
KVKVIGSPNTPEG
SPPRVKSPEPRVK
SSLRYSSPPAHVK
LILSACSPYFREY
PGIRGPSPSPVGS
RFTKVYSPAHTEE
QFSGPASPSKKPG
ETVPATSPIPASP
SPGVVSSPPPQQS
GPVREVSPAPATQ
PTPEPQSPTEPPA
KGPGLPSPTKEVD
PAAQKTSPIPESS
GGPRNLSPPLTPA
PRSARSSPPPLSG
VKNTADSPTGEAA
TNPREHSPSESEK
IIVNSHSPASSAA
SSQSSKSPSLSSK
RVSDQNSPVLPKK
GPAEAPSPRGSPR
SSPSSPSPTLGRQ
VSGSLKSPVPRSD
SKHKSESPCESQY
TAADVVSPGANSV
SYYFGLSPEERRE
GGGHPDSPGLPAP
VGVAEGSPVSRRR
PLAQPESPTASAG
TSATQSSPAPGQS
KLGASNSPGQPNS
VENRSESPIPVAD
AATAPPSPGPAQP
APTAALSPEPQDS
SPRRSSSPDKFKR
RPRTVSSPIPYTP
PVSAPPSPRDISM
VVPFQESPKYAAR
FEPRYPSPPLKPA
EKYPKTSPLPTPV
APGEQESPKNPEI
PRGPSHSPEKESA
AQEENRSPSGSQS
PVEISPSPPGQLN
RVKSTYSPLPLLK
AARRPHSPDPGAQ
LCSPRASPDASSN
GSSQLCSPRASPD
SCNAPGSPEGSSQ
GGSRSLSPMVIMC
AGLRRESPQTLKL
GRLLNSSPEGTVL
GIRVYYSPPVARR
EEGSKASPSPSPS
DVREGHSPLSKSS
AVREKISPVSLPQ
IKERPQSPGKQES
CKKRSQSPEDLSG
KRISSKSPQPEEK
SSKRHVSPEPVKM
YREPKSSPTQSLS
PGAARESPKANGQ
LDERPSSPIPLLP
PNNISFSPKAPEM
HLLQAPSPPRTSG
HGLEPASPRHSDN
GATPLASPGLQPP
TMAAPGSPSLSHR
QSPEATSPRSPGV
GQYENQSPEATSP
AAFRQGSPTPALP
VVTTPGSPSLGRH
QSFHPKSPASSTF
RSPGVRSPVQCVS
PRQQERSPLQSLA
EGTSPSSPPHSVA
GTNTPPSPGFGRR
EATSPRSPGVRSP
SSTFLPSPHSSAG
TAPAALSPQEKKE
GGSMRVSPSIQPQ
AQPAHRSPADSLS
ARKRKPSPEPEGE
CFPEGRSPTGAQP
PELNRQSPNPRRA
NSLSTLSPKSKLG
EDNKAASPGPPGL
ESAPSLSPVKELN
DSELSVSPKRNSI
TPLNSESPVRTPV
SSISQVSPERGIP
ESEGKSSPALAAP
EREKTGSPTPSNQ
FKKLDESPVLKPE
RCEENTSPTLDAV
SNQLNDSPTFKKL
KRARGISPIVFDR
ESHHGGSPIHWVL
CEVGHMSPRKNEE
TTVEQSSPSGKLK
EDSQVFSPKKGQK
NTEEPSSPVRKES
KGPRTPSPPPPIL
QESRSLSPSHLTE
IIQKEVSPEVVRS
PDGRVLSPLIIKS
VPGEPASPISQRL
ATRLQESPAPEKG
SSHSHRSPPGSAR
TPQAPQSPRRPPH
FAEAIHSPQVAGV
KILGQQSPEKKLR
PELLASSPPDNKS
PTKRNSSPPPSPN
DDSLDLSPQGRLS
TAPTKTSPRNAKK
PRRPKYSPPRDDD
TKKRSESPPAELP
RTEEPVSPLTAYQ
VRIVEYSPPSAPR
SPPPDGSPAATPE
EPASGASPGATIP
KGNKSPSPPPDGS
PEKGNKSPSPPPD
APPPAPSPPPAPA
ASSAKTSPAKQQA
FAPEPSSPGAARA
SRSPVDSPVPASM
AAERSRSPVDSPV
SLSRHSSPTEERD
QPSRPGSPTGPPE
QKGAEPSPGGTPQ
RTGATASPSAEAL
KICTPSSPSESKR
SAPAKESPKKGAH
SSASLSSPALAKG
HLLSGKSPKKSAE
PRASAVSPEKAPM
SVNRNSSPAVPAP
NATTDSSPPREHG
PTPATTSPGEKGE
GDGGDNSPSNVVS
motif: xxxRxx_S79.9_xxxxxx score: 11.83 fg: 134/1062 bg: 60/1062 fg/bg: 2.2 unadjusted p -value: 1.4e-008 tests: 166 adjusted p -value: 2.3e-006 Show/Hide Motif Occurrences
DMARRCSVTSVES
ARKRRMSREPTLD
LTVRNVSADDDAV
LVVRKASLKDSGE
PFARAKSLPPSPV
VPGRSHSLDNPPV
FVRRLRSKEATEG
FLLRPTSIRVREG
ATTRKFSLESRGG
KVKRGSSITFSVK
VEWRKGSLQLFPC
LLVRGPSEPEEKP
FCIRSVSEAGVGE
GIKREDSRGSLIP
LLKRSCSVPLPSN
KPSRRSSQPSPTT
VLDRVASVVDSVQ
HQNRLSSVTEDED
QQVRTGSTSSKED
SQYRALSVLGCGH
IFERIQSQTVGQG
KEVRKDSVLLVWE
EAQRKGSDQWTHI
DASRKKSEWEEVT
SEIRKDSCYLTWK
KKLRPGSGGEKPP
VWFRNDSELHESW
VAGRNRSSVTLYV
QPRRPASEPHVVP
LLRRRRSLSPTYI
VTDRADSCEFTVT
VTIRAGSDLVLDA
VYVRAGSNLKVDI
SVNRKDSGDYTIT
DVTRKDSGYYSLT
CWNRPDSDGGSEI
VEKRDTSTTAWQI
ISCREPSYTPGPP
IEKRETSRLSWTQ
VDVRHDSVSLTWT
YTERTKSTITLDW
ATKRTWSVVSHKC
YRRRRRSLGDMSD
KTDRSLSGQYSCT
VSPRSLSRSPIRM
ECARVKSLDAVIT
LNVRYQSNATLVC
KLQRYNSYDINRD
SLTRSHSVGGPLQ
HVTRTHSFENVNC
GKHRPSSLNLDSA
NRFRERSLSVPTD
FRERSLSVPTDSG
TFQRTISAQDTLA
SSTRPPSVTRGGI
IPKRKLSSIGIQV
CLARSASTESGFH
RMPRSNSQENVEA
RLEREDSSEEEEE
LRPRTVSSPIPYT
GRPRKASAEDLSA
GLSRSVSQETETE
QCPRGPSHSPEKE
RLQRRNSNPSTES
TRMRTASELLLDR
RHCRSLSEPEELA
TAKRISSKSPQPE
KKTRKISQTAQTY
LRRRAASDGQYEN
GLFRSQSFPDVEP
PLIRWDSYDNFSG
THKRPASVSSAAA
VPTRADSIEETAT
CCGRKGSTEALSG
AVTRSASSTSSGS
LPSRRSSLSSHSH
SLTRAPSLTSDSE
LRHRSSSEITLSE
LPLRHRSSSEITL
SPFRTPSRLSDGL
NLERHVSTLNIQL
KLERLESEEDSIG
VRRRSESSGNLPS
RQVRRRSESSGNL
IKDRRLSEEACPG
SQKRKLSGDLEAG
QLFRRRSAGELPG
motif: xxxxxx_S79.9_Dxxxxx score: 10.62 fg: 85/1062 bg: 50/1062 fg/bg: 1.7 unadjusted p -value: 1.2e-003 tests: 158 adjusted p -value: 1.7e-001 Show/Hide Motif Occurrences
RAAHQESDNENEI
QVPGGDSDEETKT
EQEGTESDEGQLP
VKDAALSDAGEVV
DLSHTSSDDESRA
AAVDDESDYQTEY
GETLEGSDAEENL
QRSKFDSDEEDED
PPIELSSDEEYYV
QTVGQGSDAHFRV
KKPELPSDEEVPE
RFVKKLSDASTLI
TFVKKLSDHSVEP
PTRLEPSDITKDA
IIDVQLSDAGEYT
LQSISPSDAGEYT
GAISKPSDSTGPI
FTQVDSSDSGEYI
SDPSDSSDPQVAK
AILQGLSDQKVCE
TGLTENSDYQYRV
IPQSRRSDTGLYS
ITDSKRSDTGTYI
LKKALKSDIGQYT
AQNEVGSDACVCA
ERKATDSDVWHKL
ATKTPVSDLRCKV
ATNDVGSDTCVGS
ATNEVGSDTCACT
GVQISFSDGRARL
CKDIKASDITKSS
PKDLKVSDITRGS
AWKPPASDGGAKI
AENSSGSDTQKIK
QWHEPVSDGGSKV
TWTEPASDGGSKV
IVPASPSDSGEWT
IFITPLSDVKVFE
EKCPRGSDKWVAC
ISETRHSDAGEYT
KIEALPSDISIDE
IHNLSESDCGEYV
INKVDHSDVGEYT
DWKEPRSDGGSPI
SWTLPKSDGGSPI
INECVRSDSGPYP
EPVTPKSDVPIQA
RSLGDMSDEELLL
TAVVDVSDRMVPP
TVSVHVSDRVVPP
VTNDVGSDSCTTM
INRTHASDEGPYK
EGPLAVSDVTSEK
VADLKVSDVTKTS
GVPTETSDAVKAS
PRGVKVSDVSRDS
KVSIQVSDRIIPP
SNPSPSSDPIKAC
RYGVSGSDQTLTI
PDSPQHSDLAPLD
IKDVSMSDLKDFL
GRNVKNSDMHLLD
PRIHRASDPGLPA
TPESGESDKESVG
KFEDEDSDDVPRK
TSDNEGSDTEVCG
SVSEINSDDELPG
PEVVGESDSEVEG
SLEKSNSDCENYL
TLPSFSSDEEDSV
LYSKYYSDSDDEL
TAFSDWSDEDVPD
KEFGDGSDENEVE
SRASHYSDQLAPR
FTREFDSDSLRHY
RTPSRLSDGLVPS
LHGGFDSDCSEDG
ESGTEESDEEPTP
GVVGKSSDGEDEQ
IGPELDSDEDDDE
EEEFSDSDEEGEG
motif: xxxxxx_T79.9_Pxxxxx score: 9.77 fg: 69/296 bg: 31/296 fg/bg: 2.2 unadjusted p -value: 2.1e-005 tests: 83 adjusted p -value: 1.7e-003 Show/Hide Motif Occurrences
STSSSSTPPQPRD
DESRAGTPSLVTY
SDEETKTPSASPR
ESVVVSTPVQEFY
RTSPSLTPWEEVE
SEAEPGTPVQWLK
ASLMTKTPSFETA
RNAAVLTPSENVE
EEWKPVTPDATSD
SPVPSPTPAQPQK
EGGDPPTPEPLKV
PVDPPGTPDYIDV
PFKGRPTPEITWS
PQKPSRTPVQEEI
PALVPGTPKAEER
PKKPEATPVPVPE
GEALLQTPECEIK
WVKLNKTPIPQTK
TPGPPSTPWVSNV
TETVPLTPVAQEA
AKNAFVTPGPPSI
AERRSPTPERTRP
RPGPPSTPEASAI
RPARRRTPSPDYD
GQLPHKTPPRIPP
PKSRSPTPPSIAA
PPGPPGTPKVVHA
NRYGKSTPLDSKP
PTIETKTPILAIN
VIGSPNTPEGPLE
AVVEEKTPEALPK
VPGPPGTPFVTLA
VPGPPGTPQVTAV
NENGEGTPSEIVV
RVTGIPTPVVKFY
EPREIVTPVTVQD
EPGPPGTPFVTAI
EPGPPGTPFVTSI
CREPSYTPGPPSA
TIGRKETPPVEER
EAPPEPTPKERKV
ADRRAWTPVTYTV
EISEPVTPKSDVP
PCDPPGTPEAIIV
DEWTTCTPPSGLQ
PIEPAPTPIAAPV
RHVRAPTPSPVRS
WVRATKTPVSDLR
AVKASETPGPVVD
ECTVSGTPELSTK
FKATKLTPNKEYI
PYVPCPTPPRKHL
PQPPPSTPATTEI
TTEITTTPERPRR
ESGRSATPVNCDQ
EIKHVETPLCFQK
AGHGPCTPPPAPA
GTAKEGTPGGTAQ
RCTAGATPLASPG
TSDPSRTPEEEPL
RTVGTNTPPSPGF
PSTAPGTPTLRKR
KKKGPRTPSPPPP
IESPFRTPSRLSD
DGSPAATPEIRVN
VTPKTVTPASSAK
VCEVSVTPKTVTP
NAYNGETPTEKLP
PGHVSPTPATTSP
DEBUGGING INFORMATION
Command line:
momo motifx -oc /media/bayegy/disk1/bioin3/phop/output/test/outdir/Result/2.Modification_motifs/ --verbosity 1 --sequence-column 'Modified sequence' --width 13 --protein-database /media/bayegy/disk1/bioin3/phop/input/test/abundance/all_sample.fa --eliminate-repeats 13 --min-occurrences 20 --score-threshold 1.0E-6 /media/bayegy/disk1/bioin3/phop/input/test/abundance/peptide_label.txt
PARAMETERS:
algorithm: motif-x
post-translationally modified peptide filenames:
file 1: /media/bayegy/disk1/bioin3/phop/input/test/abundance/peptide_label.txt
PTM filetype: PSM
modified peptide column: 'Modified sequence'
protein database filename: /media/bayegy/disk1/bioin3/phop/input/test/abundance/all_sample.fa
protein database format: FASTA
motif width: 13
filter: false
remove unknowns: true
eliminate repeats: true
eliminate repeat width: 13
min occurrences: 20
single motif per mass: false
hash fasta: false
hash fasta width: 0
score threshold: 1e-06
binomial p-value calculations: accurate
This information can be useful in the event you wish to report a
problem with the MoMo software.
Go to top